Figure 2.
Structural basis of SARS-CoV-2 E recognition by BRD4 BDs
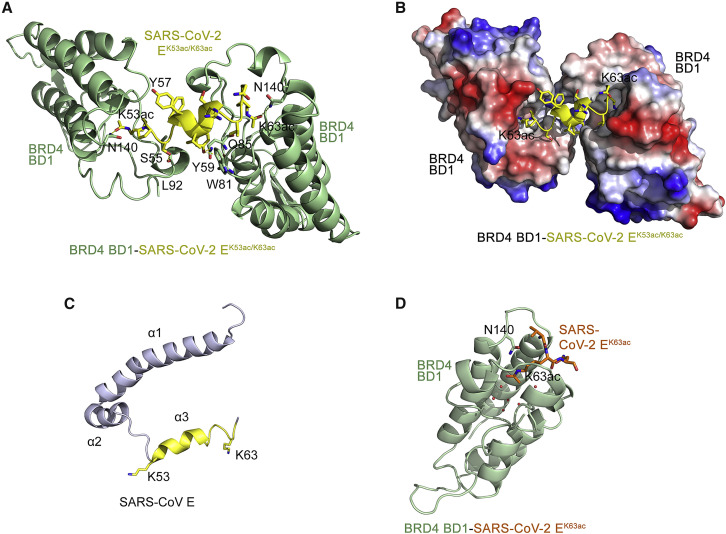
(A) A ribbon diagram of the crystal structure of BRD4 BD1 (green) in complex with the SARS-CoV-2 EK53ac/K63ac peptide (aa 53–64 of SARS-CoV-2 E, yellow). Yellow dashed lines represent hydrogen bonds.
(B) Electrostatic surface potential of BRD4 BD1 in complex with SARS-CoV-2 EK53ac/K63ac (yellow ribbon), with blue and red colors representing positive and negative charges, respectively.
(C) A ribbon diagram of the NMR structure of the homologous E protein from SARS-CoV (PDB ID: 5X29). An exposed C-terminal α-helix 3, consisting of residues K53-K63, is yellow.
(D) Crystal structure of BRD4 BD1 (green) in complex with the SARS-CoV-2 EK63ac peptide (aa 60–68 of SARS-CoV-2 E, orange). Red spheres represent water molecules. See also Figure S3.