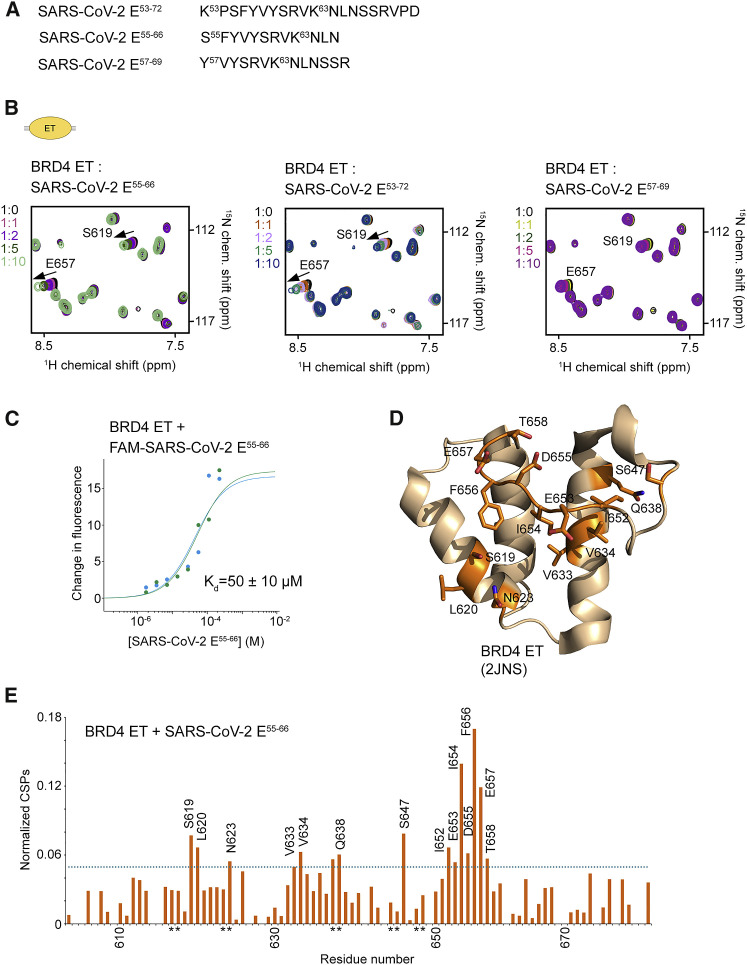
Figure 3.
The ET domain of BRD4 is an additional target of SARS-CoV-2 E
(A) The SARS-CoV-2 E peptides tested.
(B) Overlayed 1H,15N HSQC spectra of the 15N-labeled BRD4 ET domain recorded before and after gradual addition of the indicated SARS-CoV-2 E peptides. The spectra are color-coded according to the protein:peptide molar ratio.
(C) Representative MST binding curves for the interaction of the BRD4 ET domain with the FAM-SARS-CoV-2 E55-66 peptide. Data are representative of two experiments, and error represents SEM.
(D and E) Analysis of CSPs in the BRD4 ET domain induced by SARS-CoV-2 E55-66. Residues of the BRD4 ET domain that exhibited large chemical shift perturbations (greater than the average plus 1 SD) upon addition of SARS-CoV-2 E55-66 are mapped onto the BRD4 ET structure (2JNS), labeled, and colored orange in (D). Histogram of normalized CSPs in 1H,15N HSQC spectra of BRD4 ET induced by 10-fold molar excess of SARS-CoV-2 E55-66 as a function of residue is shown in (E). The dotted line indicates average chemical shift change plus 1 SD. Side chain amide resonances are indicated by asterisk.