Abstract
Tuberculosis (TB), caused by Mycobacterium tuberculosis (M. tb), is a leading cause of death due to infectious disease. TB is not traditionally associated with biofilms, but M. tb biofilms are linked with drug and immune tolerance and there is increasing recognition of their contribution to the recalcitrance of TB infections. Here, we used M. tb experimental evolution to investigate this complex phenotype and identify candidate loci controlling biofilm formation. We identified novel candidate loci, adding to our understanding of the genetic architecture underlying M. tb biofilm development. Under selective pressure to grow as a biofilm, regulatory mutations rapidly swept to fixation and were associated with changes in multiple traits, including extracellular matrix production, cell size, and growth rate. Genetic and phenotypic paths to enhanced biofilm growth varied according to the genetic background of the parent strain, suggesting that epistatic interactions are important in M. tb adaptation to changing environments.
Research organism: Other
eLife digest
In many environments, bacteria live together in structures called biofilms. Cells in biofilms coordinate with each other to protect the group and allow it to survive difficult conditions. Mycobacterium tuberculosis, the bacterium that causes tuberculosis, forms biofilms when it infects the human body. Biofilms make the infection a lot more difficult to treat, which may be one of the reasons why tuberculosis is the deadliest bacterial infection in the world.
Bacteria evolve rapidly over the course of a single infection, but bacteria forming biofilms evolve differently to bacteria living alone. This evolution happens through mutations to the bacterial DNA, which can be small (a single base in a DNA sequence changes to a different base) or larger changes (such as the deletion or insertion of several bases).
Smith, Youngblom et al. studied the evolution of tuberculosis growing in biofilms in the lab. As the bacteria evolved, they tended to form thicker biofilms, an effect linked to 14 mutations involving single base DNA changes and four larger ones. Most of the changes were in regulatory regions of DNA, which control whether genes are ‘read’ by cells to produce proteins. These regions often change more though evolution than regions coding for proteins, because they have a coordinated effect on a group of related genes rather than randomly altering individual genes. Smith, Youngblom et al. also showed that biofilms made from different strains of tuberculosis evolved in different ways.
Smith Youngblom et al.’s findings provide more information regarding how bacteria adapt to living in biofilms, which may reveal new ways to control them. This could have applications in water treatment, food production and healthcare. Learning how to treat bacteria growing in biofilms could also improve the outcomes for patients infected with tuberculosis.
Introduction
In 2019, an estimated 10 million people fell ill due to tuberculosis (TB), and one quarter of the world’s population is estimated to be infected with its causative agent Mycobacterium tuberculosis (M. tb) (Global Tuberculosis Report, 2019, WHO). New strategies for diagnosis, treatment, and control of TB are urgently needed. From an evolutionary perspective, M. tb stands out among bacterial pathogens for its strict association with human hosts, limited genetic diversity, and clonal evolution (Eldholm and Balloux, 2016). We might expect these features to constrain adaptation of M. tb, yet TB remains a challenging infection to treat due to the bacterium’s ability to persist in the face of antibiotic and immune pressure and to acquire novel drug resistances. In order to better treat and control TB, we need to understand the sources of M. tb’s robustness and to identify its vulnerabilities. Experimental evolution is a powerful tool for illuminating these strengths and vulnerabilities and has led to important insights into the fundamental processes guiding microbial adaptation.
Biofilms are increasingly recognized as a relevant growth form for bacteria in their natural environments (Costerton et al., 1999). TB is not traditionally thought of as a biofilm infection. However, M. tb cells spontaneously aggregate and secrete extracellular matrix (ECM) when grown in vitro (Dubos and Davis, 1946; Bacon et al., 2014), suggesting they are naturally inclined to grow as biofilms. Autopsy studies have long identified M. tb aggregates in human tissues during TB infection (Canetti, 1956; Nyka, 1977; Nyka, 1967; Nyka, 1963; Nyka and O’Neill, 1970). More recent research has demonstrated specific biomarkers of M. tb biofilms in human autopsy specimens and animal models of TB (Chakraborty et al., 2021). The presence of M. tb biofilms during TB infection is of major practical significance as growth within a biofilm allows M. tb cells to survive otherwise lethal concentrations of antibiotics and to evade immune responses (Ojha et al., 2008; Ackart et al., 2014; Trivedi et al., 2016; Chakraborty et al., 2021). Identifying the mechanisms of biofilm development by M. tb can thus aid the development of new, more effective therapies for TB (Wang et al., 2013; Ackart et al., 2014; Richards et al., 2019; Chakraborty et al., 2021).
The genetic determinants of M. tb biofilm formation have been investigated with candidate gene approaches and phenotypic characterization of knockout, knockdown, and overexpression mutants (Ojha et al., 2008; Pang et al., 2012; Sambandan et al., 2013; Wolff et al., 2015; Rastogi et al., 2017; Yang et al., 2017; Richards et al., 2019; Hegde, 2019; Bharti et al., 2021; Chakraborty et al., 2021). Here, we use a complementary approach based on serial passaging of M. tb clinical isolates under selective pressure to grow as a biofilm. This approach has the advantage of maintaining the integrity of complex networks of genes and their regulators while enabling discovery of subtle genetic changes with an impact on biofilm phenotype. It is also unbiased with respect to the choice of candidate loci. During experimental evolution of six closely related M. tb isolates passaged over months to years we found: (1) rapid adaptation in response to selection imposed in our system, with development of more robust biofilm growth in all strains, (2) changes in a range of M. tb phenotypes in association with few, presumably pleiotropic mutations, (3) predominant impacts on gene dosage among mutations that emerged during the experiment, (4) implication of M. tb loci not previously known to be involved in biofilm development, and (5) apparent effects of strain genetic background in shaping the adaptive path to the phenotype under selection.
Results
Sample
Pellicles are a specific type of biofilm in which bacteria form aggregates at air–liquid interfaces (Kobayashi, 2007); M. tb has been shown to form pellicle biofilms in vitro (Kerns et al., 2014). For our study, we chose six closely related isolates of M. tb, from three sub-clades of the Euro-American lineage L4 (4.9, 4.4.1.2, and 4.4.1.1 shown in Figure 1). The study design enabled us to identify the impacts of genetic background over multiple scales, including comparisons between sub-lineages (4.9 and 4.4), sub-sub-lineages (4.4.1.2 and 4.4.1.1), and individual strains. In addition, the strain selection encompassed a variety of biofilm phenotypes such that we could identify impacts of ancestral phenotype on bacterial adaptation in our passaging system. The strains were grown as pellicles following a published protocol (Kulka et al., 2012). We passaged these six populations in pellicle form every 5–7 weeks as described in the Methods section. Each pellicle population was passaged at least eight times over a period of 2 years (Figure 1—figure supplement 1; Supplementary file 1). To investigate the specificity of adaptations observed during pellicle passaging, we also passaged the six strains in planktonic culture: we passaged three independently evolving populations per strain, and each population was passaged four times.
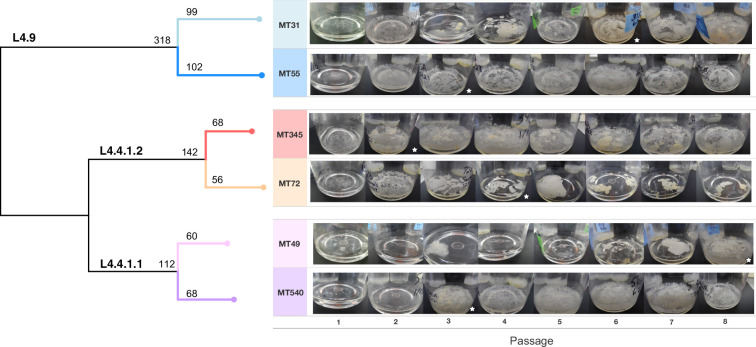
Figure 1. Whole genome sequence phylogeny of ancestral populations used for passaging and photos of pellicles throughout the experiment.
Populations fall into three clades with single-nucleotide polymorphism distances given for each sub-lineage and strain. Pellicle photos show rapid change in phenotype, followed by stabilization through the rest of the experiment. The star indicates the passage at which point we determined the pellicle phenotype to be stabilized for each population. Photos of pellicles were taken after 5–7 weeks of growth.
Figure 1—figure supplement 1. All photos from pellicle passaging experiment.
Phenotypic changes of pellicles
At each passage, we photographed the pellicle and described its growth according to the following criteria: proportion of liquid surface covered, presence of climbing (attachment to and growth up the sides of the flask), thickness of growth, and continuity of growth (versus discontinuous patches). Although the M. tb strains were closely related (i.e. separated by 100 s of single-nucleotide polymorphisms [SNPs]), differences in biofilm phenotype were evident prior to passaging (Figure 1). During the initial few passages, phenotypes changed for all strains and then stabilized between passages 2 and 8 (Figure 1). We performed extended passaging for four strains, which were carried out to passage 16 or 20; we did not observe any further phenotypic changes during this extended passaging (Figure 1—figure supplement 1).
Over the course of the experiment, all populations evolved more robust biofilms characterized by an increase in surface coverage and thicker, reticulating growth (Figure 1). MT72 was an exception, as it developed a typical confluent biofilm after two passages, but then evolved discontinuous, thick growth covering only a small fraction of the surface. This contrasts with the other evolved pellicles, which cover the entire surface of the liquid, climb up the side of the flask, and are more confluent (Figure 1).
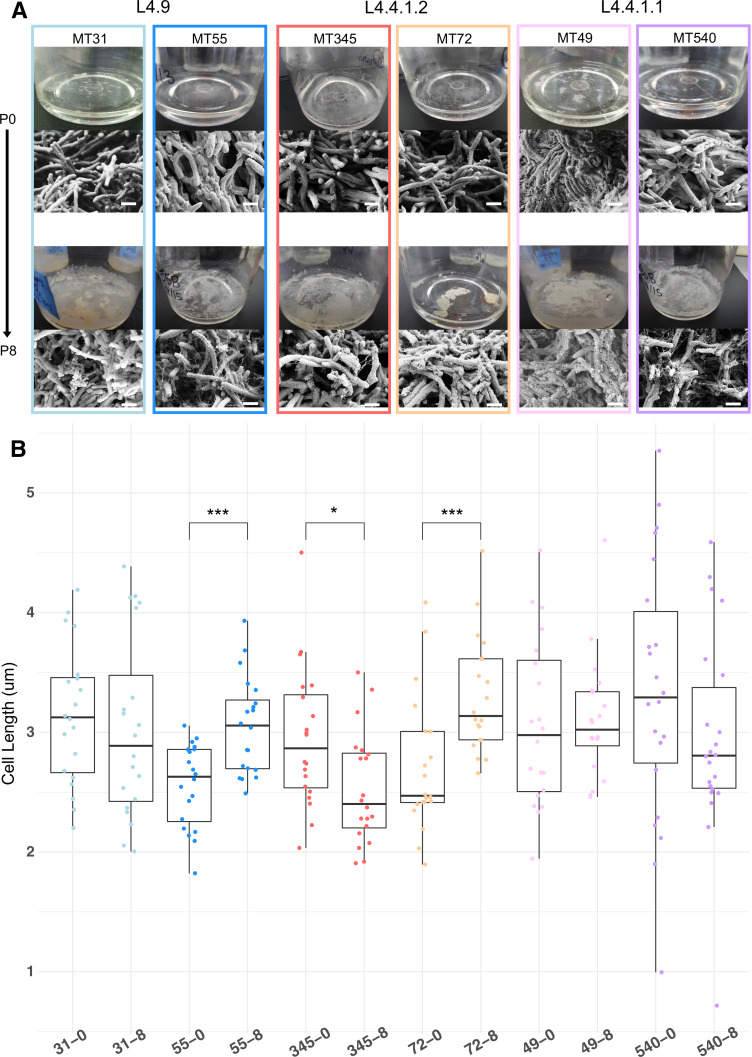
To further characterize M. tb biofilm phenotypes, we performed SEM of biofilm samples from ancestral strains and paired evolved strains that had undergone eight rounds of passaging. The appearance of ECM was variable among strains at baseline, but all strains exhibited increased production of ECM after serial passaging (Figure 2). We observed changes in ECM appearance including increased webbing (MT55 and MT540) and increased production of globules (MT31, MT345, MT72, and MT49) (Figure 2). Bacterial cell shape has been documented to have direct fitness consequences in many species (Yang et al., 2016). We measured bacilli length of ancestral and evolved strains and found that three strains (MT55, MT72, and MT345) had significant changes in cell length (Figure 2). The direction of the change varied among strains: MT55 and MT72 evolved longer bacilli whereas MT345 evolved shorter bacilli (Figure 2).
Figure 2. Phenotypic changes in evolved bacterial populations: cell size & matrix production.
(A) SEM images of ancestral and evolved biofilms show changes in extracellular matrix and (B) cell length. SEMs were taken after 5 weeks of biofilm growth, shown alongside photo of pellicle from that passage. Cell lengths were measured across two biological replicates for each strain, except MT540-0 for which only one replicate was available. Each dot represents a single cell length measurement. Significant differences (Kruskal–Wallis p-value=3.43e-05) in cell length between ancestral and evolved pairs shown across the top of panel B (Mann-Whitney U test with Benjamini-Hochberg correction). Scale bars in lower right side of SEM images = 1 μm. p-value legend: *p<0.05, **p<0.01, ***p<0.001.
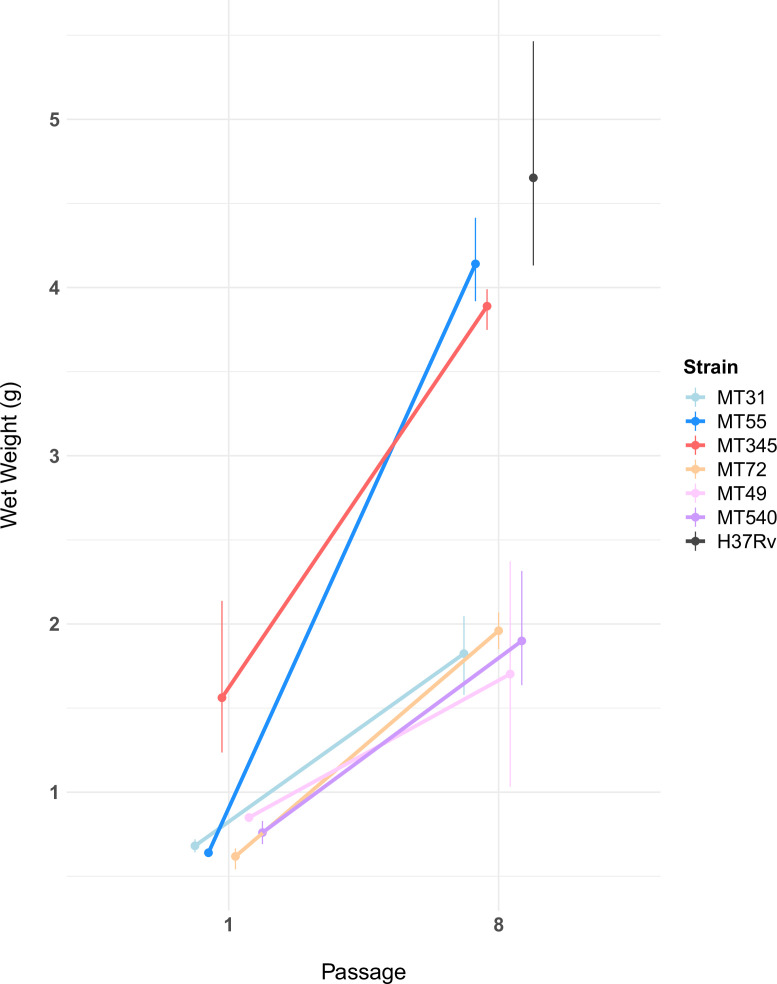
Wet weights
To obtain another quantitative measure of phenotypic adaptation to biofilm growth, we developed a protocol for measuring the wet weight of a pellicle biofilm. Wet weights of ancestral biofilms were similar except for MT345, which was heavier in keeping with its confluent morphotype (Figures 1 and 3). We observed a significant (Mann-Whitney U test, p-value=1.5e-06) increase in wet weight for all populations after eight passages (Figure 3). Of note, we observed a substantial increase in wet weight in MT345, which formed a robust pellicle at baseline, suggesting that the potential to increase pellicle biomass remains among M. tb isolates that form robust biofilms at baseline. Indeed, MT345 along with MT55 had particularly dramatic increases in pellicle biomass (an average of 2.3 g and 3.5 g, respectively) with evolved isolates approaching the phenotype of lab-adapted strain H37Rv (Figure 3). We also observed a trend towards increased variability of wet weights between replicate cultures of the evolved biofilms (Figure 3).
Figure 3. Pellicle wet weights measured after 5 weeks of growth, for ancestral populations and populations after eight passages (n=2–3 per experimental strain, n=5 for H37Rv, see Figure 3—source data 1).
Pellicle wet weights increased for all experimental populations following passaging (Mann-Whitney U test, p-value=1.5e-06). The magnitude of this change varied, with two genetic backgrounds (MT55 & MT345) showing relatively dramatic increases in pellicle weight, approaching the phenotype of lab-adapted strain H37Rv. We also observed a trend towards increased variability in wet weights after passaging as a pellicle. Error bars represent range of wet weights across replicates.
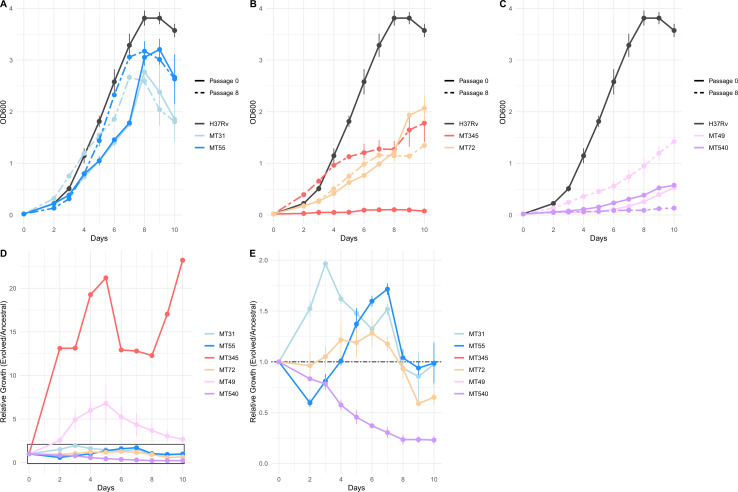
Planktonic growth trade-offs during pellicle adaptation
To investigate whether adaptation to pellicle growth involved trade-offs with fitness in planktonic culture, we compared planktonic growth rates of ancestral and pellicle-evolved populations. Some evolved strains grew faster in planktonic culture following pellicle passaging (MT31, MT345, and MT49) whereas other strains exhibited similar growth rates – if slightly different kinetics – when compared with their ancestral strains (MT55, and MT72; Figure 4). One evolved strain (MT540) had a slower growth rate than its ancestor (Figure 4C; E).
Figure 4. Impacts of pellicle passaging on planktonic growth rate vary by genetic background.
(A, B, C) Planktonic growth and (D, E) relative fitness curves comparing the growth of strains after eight pellicle passages to the ancestral strains. Growth curves were performed in triplicate, and the mean OD600 value is plotted with error bars representing ±1 SD. Relative fitness was calculated at each timepoint as OD600 Passage 8 / OD600 Passage 0. Panel E shows boxed region from panel D in more detail.
Candidate loci involved in biofilm formation
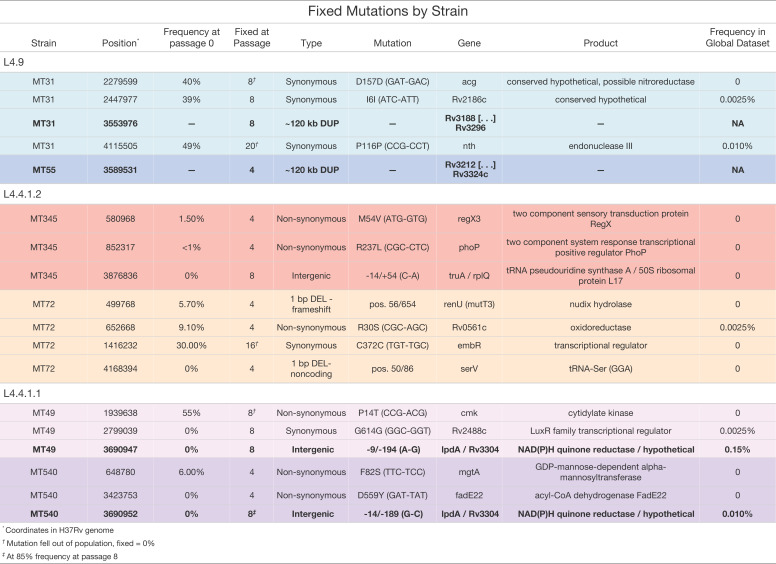
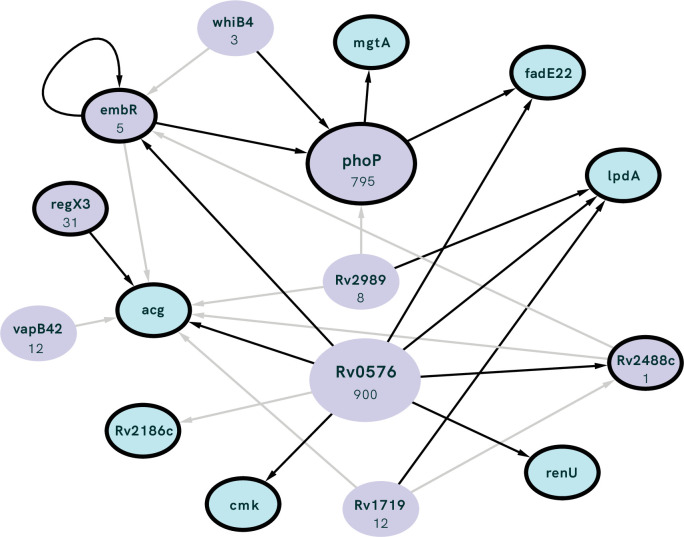
Genomic DNA (gDNA) was extracted from whole biofilm populations for (pooled) sequencing every four passages (except MT31 and MT49 which were first sequenced at passage 8; Supplementary file 1). To identify loci potentially responsible for the observed changes in pellicle phenotype, we identified mutations that rose to fixation (variant at 100% in the population) or disappeared (variant at 0% and reference allele at 100%) over the course of our experiment. After filtering and manual curation of the results (see Methods), we identified 14 SNPs, 2 duplications, and 2 deletions that met these criteria (Figure 5). The starting frequency of these alleles varied from 0 to 55% and fixation occurred by the eighth passage for all but two of these variants (position 4,115,505 in MT31 and position 1,416,232 in MT72; Figure 5). Candidate genes are annotated with a range of functions with regulation (regX3, phoP, embR, and Rv2488c) and lipid metabolism (Rv2186c, mgtA, and fadE22) being the most common (Figure 5). A search for these mutations within a global dataset containing ~40,000 isolates (see Methods) revealed these mutations to be exceedingly rare or completely absent from other sequenced strains of M. tb (Figure 5). The most commonly identified SNP was at position 3,690,947 upstream of lpdA (identified in our study in strain MT49), at a frequency of 0.0015 in this dataset (Figure 5). One caveat is that genes in repetitive families (PE, PPE, and PE-PGRS) were excluded from our analysis due to difficulties in accurately resolving these regions with short-read sequencing data. It is possible that variants contributing to enhanced biofilm formation lie undetected in these regions.
Figure 5. Small number of variants that became fixed (>95 or <5% frequency) throughout the course of the experiment.
Including both single nucleotide polymorphisms and insertions/deletions (INDELs) identified by Popoolation2, Breseq, and/or Pilon. Highlighted in bold are two instances of convergent adaptation we observed within the L4.9 and L4.4.1.1 sub-lineages. These mutations are exclusive to strains passaged as pellicles, see Supplementary file 2 for mutations arising in strains passaged planktonically.
We did not identify any overlap in mutations that arose following pellicle and planktonic passaging, either in specific SNPs or genes containing SNPs (Figure 5, Supplementary file 2). This suggests that the candidate variants for pellicle adaptation are unlikely to represent generalized lab adaptations.
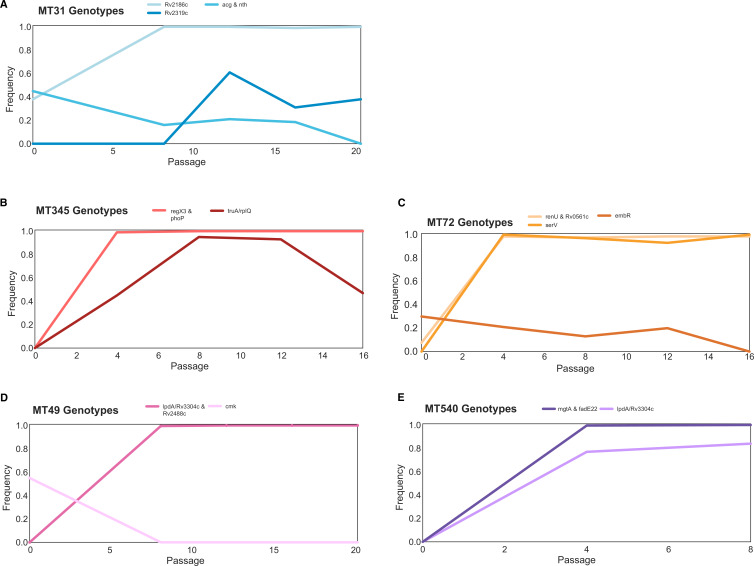
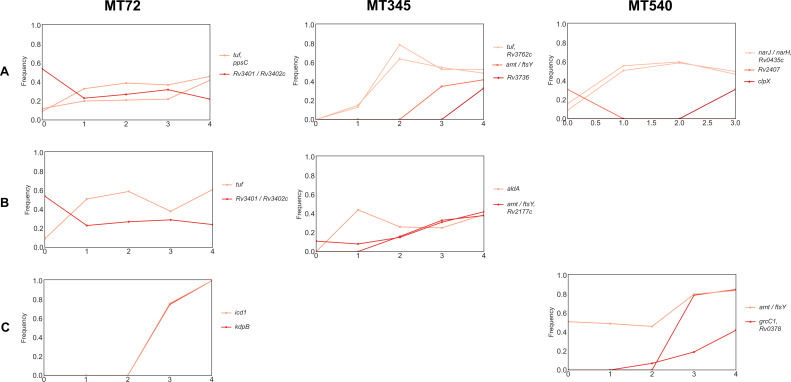
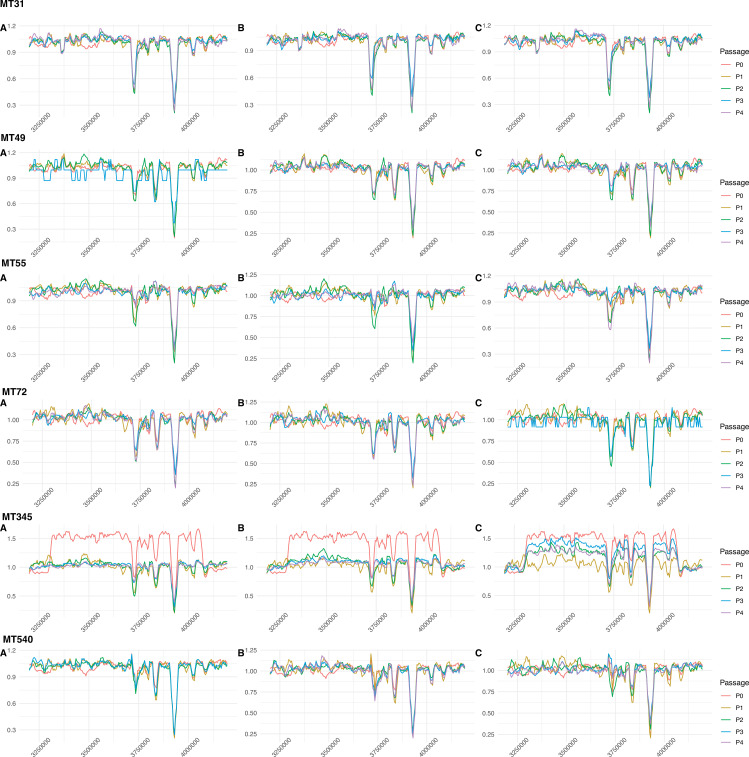
Allele frequency dynamics for all variants that changed >30% over the course of pellicle passaging are shown in Figure 6. In most populations (MT31, MT345, MT72, and MT540), we observed secondary mutations arising on the background of the first fixed mutations (Figure 6). Some secondary mutations reached high frequencies (>95%) but did not remain at high frequency for the duration of the experiment (Figure 6B). We identified a single mutation in the evolved pellicles (Figure 6A, P286L Rv2319c) with a moderate change in frequency (>30%) that did not become fixed. Allele frequency trajectories were otherwise characterized by rare variants that remained rare. Few changes in allele frequency were observed for MT55. Aside from the large duplication that reached fixation, we did not identify any other mutations in this strain with ≥30% change in frequency.
Figure 6. Trajectories of mutations that changed ≥30% over the course of the experiment show few mutations arose during passaging.
The mutations that became fixed almost always did so in the first or second sequenced passage (Supplementary file 1), and we identified very few mutations with substantial declines in frequency. Mutation frequency data were calculated with Popoolation2 and plots were made using Lolipop (see Methods).
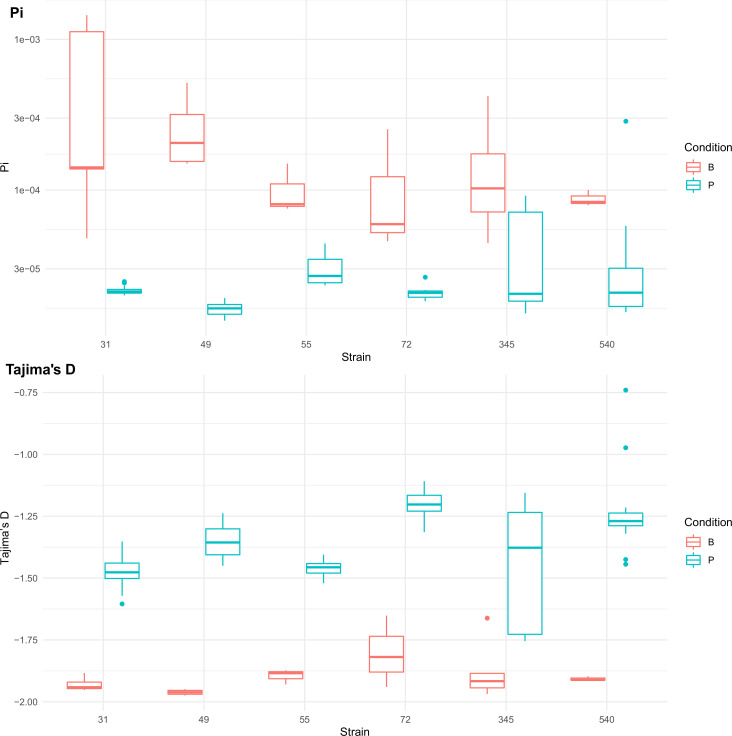
Figure 6—figure supplement 1. Population genetics statistics calculated from Pool-seq data from all strains at all sequenced passage points.
Figure 6—figure supplement 2. Trajectories of mutations that changed ≥30% over the course of planktonic passaging.
We calculated genome-wide measures of diversity (Tajima’s D and nucleotide diversity pi) for sequence data at each timepoint (Figure 6—figure supplement 1). Our values concur with previously estimated values for M. tb (O’Neill et al., 2015) and are consistent with genome-wide purifying selection (Pepperell et al., 2010; Pepperell et al., 2013). Bacterial populations within pellicles were more diverse (characterized by higher values of pi) and exhibited a stronger skew to rare variants (lower Tajima’s D) than those found in planktonically grown communities (Figure 6—figure supplement 1). This observation points to the complexity of biofilm communities and distinct demographic and/or selective forces associated with pellicle versus planktonic growth.
Convergent adaptation
We observed two instances of convergent adaptation, i.e., the same locus being subject to repeated mutation during pellicle adaptation. Interestingly, each pair of convergent mutations appeared in the same sub-lineage. A large (~120 kb) duplication fixed in populations MT31 and MT55, which both belong to sub-lineage L4.9. The second convergent mutation, an intergenic SNP upstream of NAD(P)H quinone reductase lpdA, within a transcription factor binding site (TFBS) and a non-coding RNA (ncRNA), occurred in MT49 and MT540 that both belong to sub-lineage L4.4.1.1 (Figure 5). Similar patterns were evident in the planktonically passaged populations of bacteria: we identified convergence in these experiments, with mutations arising repeatedly at the same loci (Supplementary file 2, Figure 6—figure supplement 2). We also observed a likely impact of genetic background on adaptation, with loci subject to repeated mutation in an apparently strain- or sub-lineage specific manner (Supplementary file 2, Figure 6—figure supplement 2).
Convergent adaptation affecting expression of lpdA
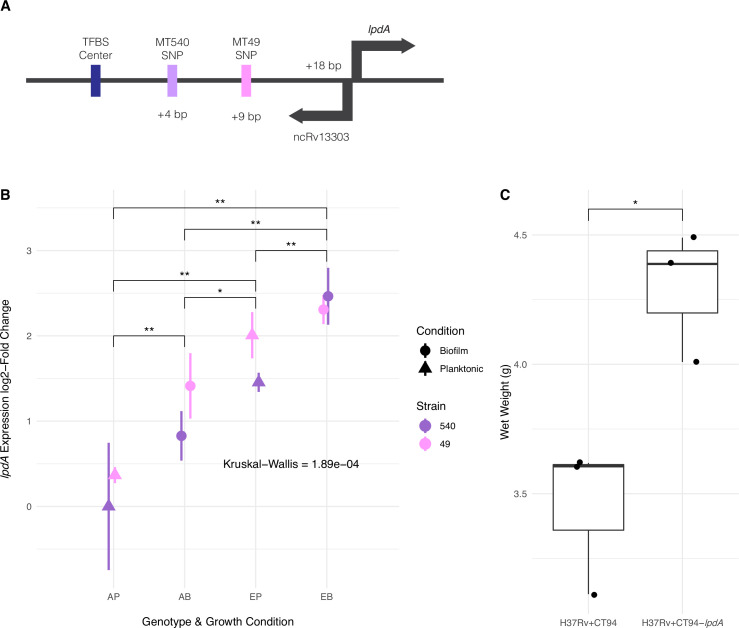
We identified convergent adaptation in evolved pellicles of strains MT49 and MT540 (both of lineage L4.4.1.1) in the region upstream of NAD(P)H quinone reductase lpdA (Figure 5). These intergenic SNPs are five base pairs apart and lie within the TFBS for Rv1719, which is known to downregulate lpdA (Rustad et al., 2014; Figure 7A). We hypothesized that an SNP within the TFBS would affect transcription factor (TF) binding and therefore expression of lpdA. We sought to quantify lpdA expression by quantitative polymerase chain reaction (qPCR) in ancestral populations and compare that to expression in evolved populations containing the SNP of interest. Evolved populations had significantly higher (p<0.05, Mann-Whitney U test) expression of lpdA (Figure 7). Additionally, we observed significantly increased expression of lpdA during biofilm growth when compared with planktonic growth, suggesting a role for this gene in biofilm formation (Figure 7). In addition to being within a TFBS affecting expression of lpdA, these SNPs also lie within an ncRNA that is expressed from the opposite strand of lpdA (Figure 7). This ncRNA was identified by RNA-sequencing (Arnvig et al., 2011; Gerrick et al., 2018) but has yet to be characterized. In order to further investigate the role of lpdA in pellicle growth, we measured pellicle phenotypes in H37Rv after introducing a second copy of the gene with an integrative, constitutive overexpression plasmid. Pellicles from bacteria with the additional copy of lpdA had significantly higher mass than those from bacteria transformed with an empty vector (Figure 7C), with no change in gross phenotype (Figure 6—figure supplement 1).
Figure 7. Adaptation to pellicle growth via increased expression of lpdA.
(A) Diagram of convergent adaptation in L4.4.1.1 strains MT540 and MT49 in the region upstream of lpdA within a transcription factor binding site (TFBS) and non-coding RNA. (B) Expression of lpdA differed significantly between sample groups (Kruskal–Wallis test, p-value=1.89e-04). Pairwise comparisons between groups (n=3, Mann-Whitney U test with Benjamini-Hochberg correction) revealed significantly increased expression in evolved (E) strains when compared with ancestral (A) strains, as well as during biofilm (B) growth when compared with planktonic (P) growth. (C) Overexpression of lpdA in H37Rv results in significantly (n=3 for each construct, Mann-Whitney U test) increased biofilm wet weights. p-value legend: *p<0.05, **p<0.01.
Figure 7—figure supplement 1. H37Rv pellicles with empty vector (left) and extra copy of lpdA (right) introduced into the chromosome using pCT94.
Convergent evolution of large duplication
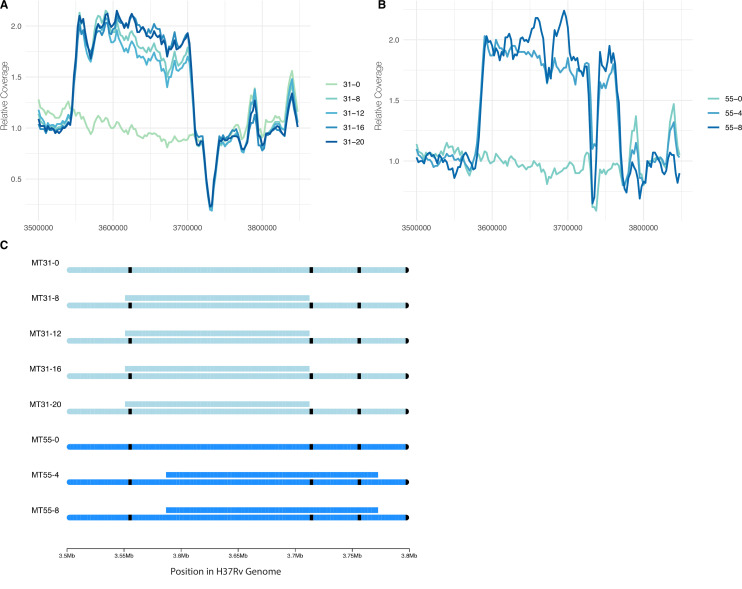
We discovered a large (~120 kb) duplication in pellicles of both strains from lineage L4.9 (MT31 and MT55, Figure 5) following pellicle passaging. The duplications were initially identified with Pilon (Walker et al., 2014), and to confirm their coordinates and absence in the ancestral populations, we made sliding window coverage plots in this region and confirmed ~2× coverage indicating the presence of a duplication (Figure 8). The coordinates of the duplication appear to be slightly different in the two populations with the duplication in MT55 starting about 3 kb after the duplication in MT31 (Figure 8). We determined that the duplication was fixed after the first sequenced passage in each population (passage 4 and 8 for MT55 and MT31, respectively; Supplementary file 1). After fixation, the duplication appears to have remained stable in both populations, as far out as 20 passages in MT31, as we do not see any reduction in coverage across or within the duplicated region at these later passages (Figure 8). This duplication did not emerge in planktonically passaged isolates of MT31 and MT55, suggesting it has some specificity for pellicle adaptation of these strains (Figure 7—figure supplement 1).
Figure 8. Convergent evolution of a large duplication in L4.9 strains MT31 and MT55, within the same genomic region flanked by mycobacterial IS6110 insertion sequences.
Sliding window coverage plots show the increase in coverage in this region for strain (A) MT31 and (B) MT55, and a chromosome plot (C) shows the duplications’ coordinates and overall stability in both populations over multiple passages along with the location of IS6110 insertion sequences in black.
Figure 8—figure supplement 1. Sliding-window plots showing relative sequencing coverage of planktonically passaged isolates (n=3 independently evolving replicates per strain).
Figure 8—figure supplement 2. Results of 100 permutations of random gene ontology (GO) term enrichment analysis (see Methods).
There are a total of 172 genes within the duplicated regions in MT31 and MT55 (metU through folD). To identify a possible functional enrichment within these genes, we performed a GO term enrichment analysis and compared our results to the results from randomly selected groups of contiguous genes from throughout the genome (see Methods). While the initial analysis identified significant enrichment of genes involved in nucleotide metabolism within the duplication (Supplementary file 3), this enrichment does not appear significant when compared with randomly selected gene sets (Figure 8—figure supplement 2).
Regulatory pathways involved in biofilm formation
Having identified two instances of convergent evolution at the level of individual loci, we investigated further for evidence of convergent evolution at the regulatory pathway level. We identified 14 genes of interest (GOI) in association with variants that rose to fixation during the passaging experiment (Figure 5). Using the MTB Network Portal v2 database of transcription factor overexpression (TFOE) and ChIP-Seq data (http://networks.systemsbiology.net/mtb/), we identified regulatory pathways that contained GOI. In Figure 9, we show the TFs that are predicted to affect the expression of multiple GOI. Of our GOI, acg is the most highly regulated, with seven TFs predicted to affect its expression. The function of acg is unknown: it is annotated as a conserved hypothetical, but according to in vivo studies may play an important role in infection (Hu and Coates, 2011; Singh et al., 2019). Additionally, we used these data to determine if GOI annotated as regulators (regX3, phoP, Rv2488c, and embR) regulated other GOI (Figure 9). We found again that acg appears to be highly regulated, and that phoP also regulates mgtA and fadE22, which are both hypothesized to have a role in cell-wall processes. Taken together these results indicate that M. tb has a highly complex and tightly regulated network and suggests that biofilm formation involves a complex network of interacting loci.
Figure 9. Interconnected regulation of our genes of interest (GOI; outlined in black) by common transcription factors (TFs; purple) with the number of genes regulated by each TF listed under gene name.
TF identified as having significant overlap with our GOI (i.e. the number of GOI regulated by the TF is higher than expected by chance) using the MTB Network Portal (see Methods). Edges between nodes indicate >0.5 log2-fold absolute change in expression in a transcription factor overexpression strain, black lines show statistically significant (p<0.05) differential expression.
Discussion
Variation within the Mycobacterium tuberculosis complex (MTBC)
It has long been recognized that the MTBC is divided into well differentiated, globally circulating lineages (Hirsh et al., 2004). As an obligate human pathogen, M. tb is subject to the vagaries of human behavior, and this behavior has had a prominent role in structuring M. tb populations (Pepperell et al., 2011; Brynildsrud et al., 2018; Liu et al., 2018; Mulholland et al., 2019; O’Neill et al., 2019). In addition to genetic variation, phenotypic differences have been described among strains of M. tb. Differences have been observed in a range of phenotypes including metabolism (Øyås et al., 2020), gene expression (Domenech et al., 2017), DNA methylation (Modlin et al., 2020), in vitro stress responses (Tizzano et al., 2021), and biofilm morphotype (Pang et al., 2012).
We have shown previously that M. tb populations are genetically sub-divided over fine scales as a result of neutral genetic drift (Pepperell et al., 2010). Here, we find that M. tb strains from within a single lineage (lineage 4, ‘Euro-American’), separated by few genetic differences (Figure 1), exhibit variation across a range of phenotypes. Prior to the imposition of selection pressures in vitro, differences among strains were evident in pellicle morphotype (Figure 1), cell length (Figure 2), pellicle wet weight (Figure 3), and planktonic growth rate (Figure 4). In some cases, phenotypic clustering occurred at the sub-lineage scale – e.g., planktonic growth curves of L4.9 isolates – and in others sub-lineage strain pairs exhibited marked differences – e.g., planktonic growth rate and pellicle wet weight of the L.4.1.1.2 strains MT345 and MT72.
Genetic and phenotypic responses to selection for biofilm growth
Regardless of phenotypic starting point, all strains of M. tb in our study showed evidence of adaptation in response to selection for pellicle growth. The gross morphology of M. tb pellicles changed over the course of the experiment (Figure 1), pellicle wet weights increased for all strains (Figure 3) and increases in the amount of ECM were evident in SEM for all strains (Figure 2). MT345 formed robust pellicles at baseline yet manifested a dramatic increase in wet weight following pellicle passaging, indicating that adaptive capacity remains even among high biofilm forming clinical isolates (Figure 3). Collectively, these results demonstrate that our method was effective in exerting selection for pellicle growth. More broadly, it shows that experimental evolutionary approaches can be applied successfully to complex phenotypes and fastidious, slow growing, and non-model organisms.
During our experiment, pellicle phenotypes stabilized between passage 2 and 8 (Figure 1); extended passaging beyond eight passages did not produce further phenotypic changes (Figure 1—figure supplement 1). Fixation of novel and previously segregating mutations occurred over the same time frame (Figure 5; Figure 6), and we did not observe any further sweeps over the course of the experiment. Within eight passages, we thus saw phenotypic and genotypic stabilization across all populations. This phenomenon is frequently observed during experimental evolution, where rapid adaptation is followed by diminishing returns after continued passaging (Barrick et al., 2009; Khan et al., 2011; Philippe et al., 2007).
We observed few mutations over the course of the experiment (Figure 5), and some of these are likely to be hitchhikers linked to advantageous alleles that swept to fixation (Figures 5 and 6). These few mutations appeared to have pleiotropic effects, as we observed changes in phenotypes other than the one under selection. ‘Off target’ phenotypes, such as planktonic growth rate and cell length, varied among strains. Experimentally evolved populations generally manifest first-pass, large-effect mutations during initial adaptation; our results suggest that these mutations in M. tb are broad in their scope and variable in their effects. The latter phenomenon may be due to a large genetic target for biofilm growth, the impacts of hitchhiking mutations, and/or genetic background of the strains.
Candidate SNPs and deletions mediating pellicle adaptation were either exceedingly rare or absent in our search of a database of almost 40,000 M. tb genomes. This suggests that the loci hit in our experiment are under strong purifying selection in M. tb’s natural environment: that is, the mutations we observed are associated with substantial fitness costs to the bacterium during human infection. This is not to suggest that biofilm growth is irrelevant to natural infection; as discussed above, M. tb biofilms and biofilm biomarkers have been identified in clinical specimens (Canetti, 1955; Nyka, 1977; Nyka, 1967; Nyka, 1963; Nyka and O’Neill, 1970; Chakraborty et al., 2021). Rather, we hypothesize that the mutations we observed are selected against because they interfere with regulatory pathways that normally constrain biofilm growth to specific environmental conditions. Regulatory constraints would limit potential drawbacks of biofilm formation, such as the metabolic costs of matrix production and immunostimulation. Another, not mutually exclusive, explanation is that fitness costs associated with mutations observed here are due to effects on off-target traits such as planktonic growth rate, cell size changes, and/or phenotypes unmeasured here. Experiments such as this one can illuminate impermissible paths in M. tb adaptation and reveal specific vulnerabilities with the potential to be exploited in TB therapeutics.
Two strains in our study evolved a large (~120 kb) duplication in the same genomic region, which fixed by passage 8 (Figures 5 and 8). The duplication persisted without evidence of decay in extended passaging in our study (up to passage 20 in MT31). Duplications in this region of the genome have been identified previously in isolates from lineages 2 and 4 (Domenech et al., 2010; Domenech et al., 2014; Weiner et al., 2012). Duplications at this site are likely facilitated by flanking IS6110 elements (Figure 8C) which can induce unequal homologous recombination. Prior research has shown that the duplication confers a fitness disadvantage in vivo (as measured in a mouse model) and can be selected during axenic culture (Domenech et al., 2014).
Here, we find this lab adaptation to be associated with enhanced pellicle formation. Interestingly, while the duplication was associated with increases in planktonic growth rate, we did not identify it in planktonically passaged populations (Figure 4, Figure 7—figure supplement 1). Domenech et al., 2014 found previously that emergence of a similar duplication was variable among strains and culture conditions. Together, these data suggest that while the duplication is observed commonly in the lab, it is selected on specific genetic backgrounds and in response to specific environmental conditions. Large-scale genome rearrangements including duplications have previously been identified as a rough adaptive ‘first draft’ followed by further refinement during experimental evolution (Lynch and Conery, 2000; Zhang, 2003). The duplication was the only mutation that fixed in MT55, which exhibited a dramatic increase in pellicle weight (Figure 3) and increased production of ECM (Figure 2) following passaging, suggesting that it is indeed this seemingly multipurpose mutation that is responsible for pellicle adaptation. The apparent impacts of the duplication were not limited to pellicle growth, as MT55 also exhibited an increase in cell length (Figure 2) and a faster rate of growth during planktonic culture, as did its sister strain MT31 also encoding the mutation (Figure 4). Taken together, our findings suggest that the production of ECM and other aspects of biofilm growth are tightly regulated during natural infection, and that first pass mutations outside of this environment result in de-repression of biofilm production. Our findings further suggest that biofilm formation is co-regulated with a range of phenotypes, consistent with prior research on biofilms, persistence and drug tolerance (Richards et al., 2019).
Epistasis
The 120 kb duplication occurred on two strains from the same sub-lineage (MT31 and MT49 from L4.9) and did not occur in any other genetic background. A similar phenomenon emerged in association with sub-lineage L4.4.1.1, where both strains underwent mutation at a TFBS upstream of lpdA, with attendant impacts on gene expression. While our strains are all very closely related, there are still distinct genetic variants for each sub-lineage that may affect the evolutionary trajectory and lead to convergent adaptation of strains within them (Supplementary file 4). We observed similar patterns in the planktonically passaged populations, for example in repeated mutations occurring in an apparently sub-lineage (tuf, L4.1.1.2) or strain-specific manner (Supplementary file 2, Figure 6—figure supplement 2). Repeated mutation at these loci provides strong support for their involvement in the trait under selection. In addition, the association of candidate adaptive mutations with specific sub-lineages suggests that M. tb strains’ genetic backgrounds played a role in shaping the adaptive path to enhanced biofilm growth.
Epistasis, which refers to the phenotypic impact of interactions among mutations at distinct loci, has been observed previously during experimental evolution of microbes (e.g. Fisher et al., 2019). Sign epistasis is a form of epistasis in which a mutation is advantageous on one genetic background and deleterious on another (Weinreich et al., 2005). The presence of sign epistasis can hinder access to a fitness optimum, and thus it represents a constraint on adaptation (Weinreich et al., 2005; Poelwijk et al., 2007). Our identification of mutations only in association with specific sub-lineages is consistent with sign epistasis, as is the observation of parallel adaptation in this small sample of strains: it suggests, for example, that mutations upstream of lpdA may be deleterious on some genetic backgrounds, and/or that alternatives are deleterious on the L.4.1.1.1 background. A careful review of the existing literature reveals additional evidence of sign epistasis in M. tb. For example, we and others have found evidence suggesting that M. tb lineage affects the mutational path to drug resistance (Mortimer et al., 2018; Farhat et al., 2019; Castro et al., 2020). Even more striking, Manson et al. identified a stereotypic order of resistance mutation acquisition in a large survey of drug-resistant M. tb isolates, a finding that was subsequently replicated (Manson et al., 2017; Ektefaie et al., 2021). This suggests that the acquisition of an initial resistance mutation alters the fitness associated with subsequent mutations, as has been observed in other settings (Silva et al., 2011). More direct evidence for sign epistasis comes from mutagenesis experiments, in which the lethality of gene knockouts has been shown to vary according to M. tb strain genetic background (Maksymiuk et al., 2015; Carey et al., 2018).
Upregulation of lpdA during biofilm growth
As noted above, we identified convergent adaptation in the region upstream of lpdA (Rv3303c) on L.4.1.1.1 backgrounds. lpdA is annotated as a NAD(P)H quinone reductase, which is known to be expressed in vivo (Deb et al., 2002) and to contribute to virulence in mice (Akhtar et al., 2006). It has not previously been linked with biofilm formation. The mechanism of its contribution to virulence in vivo is thought to be by protection against oxidative stress (Akhtar et al., 2006). The strains in our study evolved variants 5 bp apart, both of which lie within the TFBS for Rv1719 (Figure 7A). These SNPs also lie within ncRNA ncRv13303c, which was originally identified by RNA sequencing (Arnvig et al., 2011; DeJesus et al., 2017) but is yet uncharacterized. Previous work characterizing variable-number tandem repeats (VNTR) in this intergenic region have proposed a possible hairpin structure that lies within ncRv13303c and could affect its function (Zheng et al., 2008). Differences in VNTR copy number and promoter sequence in this region have been observed between different strains of mycobacteria, and even linked to differences in expression of lpdA (Akhtar et al., 2009; Pérez-Lago et al., 2013; Zheng et al., 2008).
We hypothesized that SNPs in the TFBS upstream of lpdA would affect TF binding and expression of lpdA. This was indeed the case, as evolved strains had significantly higher expression of lpdA (Figure 7B). Additionally, we found expression of this gene to be higher during biofilm growth when compared with planktonic growth. Further support for a link between increased expression of lpdA and enhanced pellicle growth comes from our data showing an increase in pellicle wet weight following introduction of a second copy of the gene via an integrative plasmid (Figure 7C). Interestingly, lpdA is within the bounds of the large duplications we identified in strains MT31 and MT55, which may affect the dosage of this gene in those evolved strains.
The two SNPs upstream of lpdA were exceedingly rare among clinical isolates of M. tb: in a search of almost 40,000 M. tb genomes, we identified the MT49 SNP in just 61 genomes and the MT540 SNP was found in 4 genomes (Figure 5). Interestingly, the MT49 mutation was shown to sweep to fixation in a deep sequencing study of serial sputum samples chronicling M. tb within host dynamics (Trauner et al., 2017). This supports the hypothesis that biofilms are important at a specific phase of TB infection, and that enhanced biofilm growth can also occur in vivo through increased expression of lpdA and genes co-regulated with it. Taken together, these observations suggest that variants at the TFBS upstream of lpdA can be transiently selected during infection but are otherwise deleterious, possibly due to fitness costs associated with biofilm growth that is timed inappropriately.
The regulatory system as a target of selection in M. tb
The L4.9 duplication and SNPs upstream of lpdA presumably act via regulation/gene dosage. Regulatory activity was also common among the other loci that accumulated mutations in our experiment, including phosphorelay signal transduction and DNA-binding activity (Figure 5). We identified fixed SNPs in four transcriptional regulators: regX3, phoP, embR, and Rv2488c (Figure 5). These genes, in turn, regulate other candidate genes identified in our study (Figure 9). regX3 and phoP belong to two-component regulatory systems (2CRS), whereas embR and Rv2488c are of the ompB and luxR families, respectively. Many of M. tb’s 2CRS have been implicated in virulence and survival in host conditions (Li et al., 2019) and this is true of regX3, which with its partner senX3 is required for virulence in mice (Li et al., 2019; Parish et al., 2003). Of note, regX3 regulates genes that are known to be involved in M. tb biofilm development (Richards et al., 2019). Similarly, while Rv2488c has not previously been directly implicated in M. tb biofilm growth, luxR regulators are involved in quorum sensing in gram-negative bacteria and have been hypothesized to play a similar role in mycobacteria (Chen and Xie, 2011; Sharma et al., 2014). Our results provide support for further investigation of quorum sensing in M. tb and its relationship to biofilm development.
Several studies have demonstrated that clinical isolates of M. tb vary in their methylation patterns, and methylome phenotypes have been linked with allelic variants in cognate methyltransferases (Modlin et al., 2020; Phelan et al., 2018; Shell et al., 2013; Zhu et al., 2016). Methyltransferase motifs are associated with regulatory elements such as promoters, and variation in methylation patterns has been associated with differences in gene expression in some cases (Chiner-Oms et al., 2019; Gomez-Gonzalez et al., 2019; Modlin et al., 2020). We did not observe any mutations at loci known to encode methyltransferase genes, but it is possible that adaptation to pellicle growth was accompanied by epigenomic changes, or that epigenomic changes occurring by an unknown mechanism contributed to the phenotypic changes we observed. Such epigenomic changes would have to be relatively stable, as the phenotypic changes we observed persisted following growth under standard planktonic conditions.
Broadly speaking, our results show that selection in our system appears to have targeted genes that regulate many other genes (hubs, e.g. phoP), and genes under regulation by many genes (e.g. acg), whose expression is likely responsive to a broad range of conditions. This reflects general features of M. tb’s regulatory network, which exhibits a high degree of connectivity between regulatory pathways (Chauhan et al., 2016; Galagan et al., 2013) as well as a hierarchical structure in which master regulators can rapidly calibrate global patterns of gene expression (Chauhan et al., 2016; Parvati Sai Arun et al., 2018). Similar findings have also been obtained in other systems, where experimentally evolved populations have shown adaptation in TFs and other global regulators, with global impacts on gene expression (Ali and Seshasayee, 2020; Conrad et al., 2010; Philippe et al., 2007; Rodríguez-Verdugo et al., 2016; Saxer et al., 2014). These mutations occur quickly to enable rapid adaptation to new environments, and it is hypothesized that secondary selection may act to refine gene expression after the initial burst of adaptation (Ali and Seshasayee, 2020; Rodríguez-Verdugo et al., 2016). Thorpe et al., 2017 previously found evidence of positive selection at promoter sites in natural populations of M. tb, suggesting that adaptation in natural as well as experimental populations is facilitated by regulatory mutations. The broad potential impacts of regulators such as regX3, phoP, embR, and Rv2488c and large-scale duplications likely helps to explain the wide range of phenotypic changes we observed. Further passaging of our isolates will reveal whether these adaptations can be further refined or whether pellicle growth remains inextricably linked to a range of phenotypes.
Of note, none of the genes identified in our study have previously been implicated in biofilm development, except for renU/mutT3. Interestingly, previous research (Wolff et al., 2015) found that renU knockouts were defective in pellicle growth, whereas, in our study, a frameshift mutation in renU was associated with enhanced biofilm growth (MT72). The novelty of our findings highlights the potential for experimental evolution to complement more traditional approaches of interrogating important pathogen phenotypes, particularly in elucidating complex interactions and the phenotypic impacts of subtle genetic changes.
From some perspectives, the adaptation of M. tb seems impossibly constrained: subject to genetic drift imposed by its host’s unpredictable behavior, subject to the limitations of a fully linked genome, and navigating a fitness landscape rendered complex by powerful epistatic interactions. Yet M. tb adapts rapidly in both natural and experimental settings. M. tb’s complex regulatory architecture exhibits many of the features that characterize robust systems – such as redundancy, modularity, and multiple feedback mechanisms (Kitano, 2004) – and we hypothesize that it is among the reasons M. tb continues to persist and thrive across the globe.
Materials and methods
Bacterial strains and growth conditions
Clinical strains were initially isolated from sputum samples. Briefly, sputum samples were de-contaminated and struck on Löwenstein–Jensen slants. M. tb growth was inoculated into Middlebrook 7H9 broth (HiMedia) containing 0.2% w/v glycerol, 10% v/v OADC supplement (oleic acid, albumin, D-glucose, and catalase; Becton Dickinson), and 0.05% w/v Tween-80 (7H9OTG) and incubated at 37°C with 5% CO2 for 3–5 weeks. Cultures were sub-cultured in 7H9OTG prior to the start of this experiment. Six clinical strains were selected for experimental passage (MT31, MT49, MT55, MT72, MT345, and MT540). The terminal culture (passage 0) of each strain was grown to an OD600 ~1 in planktonic culture and inoculated into media for biofilm growth (passage 1; Figure 10). Aliquots of passage 0 were frozen, and gDNA was isolated for sequencing. For planktonic growth, 250 μL was inoculated into 5 mL 7H9OTG, in duplicate, and incubated at 37°C with shaking, until grown to an OD600 ~1. For biofilm growth, 250 μL of planktonic culture was inoculated into 25 mL Sauton’s medium (for 1 L: 0.5 g KH2PO4, 0.5 g MgSO4, 4 g L-asparagine, 2 g citric acid, 0.05 g ammonium iron (III) citrate, 60 mL glycerol, adjust pH to 7.0 with NaOH) containing 0.1% w/v ZnSO4, in duplicate, and incubated at 37°C, with 5% CO2, without shaking for 5–7 weeks. Biofilms were grown as previously described (Kulka et al., 2012): 250 mL bottles (Corning, 430281) were incubated with a tight-fitting cap for 3 weeks, and then with a loose-fitting cap for the remainder of growth.
Figure 10. Protocol for serial passage of M. tb pellicles.
Experimental evolution of pellicles
As illustrated in Figure 10, duplicate populations of six clinical strains were grown as a biofilm. After 5–7 weeks, the more robust of the two pellicles was selected for passage. About 0.3 g (wet weight) of pellicle was inoculated into 25 mL Sauton’s in duplicate and grown as previously described. The pellicle not selected for passage was discarded. Between 8 and 20 total pellicle passages per strain were performed. Every four passages culture were frozen at –80°C and gDNA was extracted for sequencing (excluding MT31 and MT49, for whom the first passage that was frozen and had gDNA extracted was passage 8; Supplementary file 1).
Experimental evolution of planktonic cultures
Starter cultures of ancestral strains were made from frozen cryovial stocks and sequenced to get an ancestral baseline for the planktonic passages. Each strain was inoculated in triplicate to form three independently evolving populations: A, B, and C. For each passage, 25 mL of 7H9OTG was inoculated with 500 μL of culture at OD600=1. Cultures were grown at 37°C with shaking until an OD600 ~1, before freezing cryovials, extracting gDNA for sequencing, and seeding the next passage. A total of 15 of 18 of the populations were passaged four times, while 3 of 18 were only passaged three times due to the presence of contamination.
Pellicle phenotypes and wet weights
Pellicle growth was photo documented and phenotyped after 5–7 weeks of growth during the course of the experiment. Wet weights were taken after regrowing both ancestral and passage 8 strains from cryovial stocks, as described above, inoculating first into 7H9OTG and then into Sauton’s either in duplicate or triplicate. Wet weight measurements were taken after 5 weeks of pellicle growth by removing the spent media beneath the pellicle and weighing. Then, the tare weight of the empty bottle was subtracted.
Scanning electron microscopy
For SEM experiments, samples were taken from M. tb pellicle cultures after 5 weeks of growth and placed on poly-L-lysine-treated plastic coverslips (13 mm, Thermonax plastic for cell culture). Samples were then fixed overnight in a 4% formaldehyde, 2% glutaraldehyde solution in Dulbecco’s phosphate-buffered saline (-calcium, -magnesium) (DPBS) (Hyclone Laboratories Inc, Logan, UT). Following fixation, samples were washed with DPBS, then treated with 1% osmium tetroxide for 1 hr. Following osmium tetroxide treatment, samples were washed with DPBS. Next, cells underwent ethanol dehydration, which was followed by critical point drying. Following that, samples were then placed on aluminum stubs and sputter-coated with 20 nm platinum. The samples were imaged at 3 kV by a Zeiss GeminiSEM 450 SEM.
To measure cell length via SEM, SEM images taken at 10,000× were opened on ImageJ. The pixel length of the scale bar was measured with the ‘straight’ tool. Next, the image was cropped to remove any area without M. tb cells. Six frames per image were selected using the ‘unbiased frames’ macro in ImageJ. The cell length in pixels of a fully exposed M. tb cells was measured in each frame using the ‘straight’ tool. A random number generator was used to select frames for the remaining four cell lengths. After all cell lengths were measured, pixel length was converted to µm using the scale bar prior to analysis. Cell lengths were measured across two biological replicates (two independent cultures) for all strains except SK540-0, where only one replicate was available.
Planktonic growth curves
To compare rates of growth between ancestral and evolved populations, we inoculated 5 mL 7H9OTG with 250 μL thawed culture from a previously frozen cryovial for each of the ancestral strains and passage 8 of each of the pellicle evolved strains. Cultures were incubated at 37°C with shaking, until grown to an OD600~1. 15 mL fresh 7H9OTG was seeded at a starting OD600 of 0.02 in triplicate and incubated at 37°C with shaking. OD600 measurements were taken every day for 10 days (starting at day 2).
DNA extraction
gDNA for whole genome sequencing was isolated by following a modified Qiagen DNeasy Blood and Tissue Kit protocol. Briefly, cultures were pelleted and resuspended in Tris-EDTA (TE) buffer in 2 mL screw-cap tubes containing ~250 µg of 0.1 mm glass beads. The samples were heated to 80°C for 50 min to sterilize the suspensions, then frozen at –80°C for at least 1 hr to aid cellular lysis. Tubes were vortexed for 3 min for mechanical lysis. Suspensions were then incubated with 3% w/v mutanolysin (Sigma-Aldrich, M9901) for 1 hr at 37°C. Beads and cellular debris were pelleted, and supernatants were transferred to tubes for DNA purification following the protocol detailed in the Qiagen DNeasy Blood and Tissue Handbook, starting with step 4 of ‘protocol: pretreatment for gram-positive bacteria’.
Library preparation and sequencing
Library preparation was performed using a modified Nextera protocol as described by Baym et al., 2015 with a reconditioning PCR with fresh primers and polymerase for an additional five PCR cycles to minimize chimeras and a two-step bead-based size selection with target fragment size of 650 bp. Libraries were submitted to the University of Wisconsin-Madison Biotechnology Center (UWBC). Quality and quantity of the finished libraries were assessed using an Agilent DNA High Sensitivity chip (Agilent Technologies, Santa Clara, CA) and Qubit dsDNA HS Assay Kit, respectively. Libraries were standardized to 2 μM. Samples were sequenced generating paired end reads using Illumina HiSeq 2000. The majority of samples (87/101) were sequenced using the above protocol; however, for a small number of samples that needed resequencing, gDNA was submitted to the UWBC and to the Microbial Genome Sequencing Center for both library prep and sequencing using Illumina NovaSeq 6000 and Illumina NextSeq 2000, respectively.
Reference-guided assembly and variant calling
Genome assemblies were performed using an in-house reference-guided assembly pipeline (Youngblom, 2021). Briefly, raw data was checked for quality with FastQC v0.11.8 (Andrews, 2018) and trimmed using TrimGalore v0.6.4 (Krueger, 2019). Reads were mapped to the H37Rv reference genome using BWA-MEM v0.7.12 (Li, 2013), and Samtools v1.11 (Li et al., 2009) view and sort were used to process SAM and BAM files. Picard v1.183 (Broad Institute, 2022) was used to remove duplicates and add read information. Reads were locally realigned using GATK v3.5 (DePristo et al., 2011) and variants were identified using Pilon v1.16 (Walker et al., 2014). Finally, assembly quality was determined using Qualimap BamQC (Okonechnikov et al., 2016).
Lineage typing and phylogenetic tree
We used SNP-IT (Lipworth et al., 2019) to identify the lineages of our ancestral strains. To identify the sub-lineage of our ancestral strains, we obtained an alignment of known L4 isolates from a collaborator. We created an SNP alignment of the assemblies from the RGAPepPipe using Snp-sites v 2.4.1 (Page et al., 2016) and constructed a maximum likelihood phylogeny using RAxML v8.2.3 (Stamatakis, 2014). Next, we visualized the phylogeny using Dendroscope (Huson and Scornavacca, 2012) and lineage typed our isolates by comparing their location in the phylogeny to known L4 subtypes. The phylogenetic tree in Figure 1 was plotted and colored in R using ggtree (Yu et al., 2017).
Identification of fixed SNPs
To identify genetic changes occurring after repeated passaging as a pellicle or planktonic culture, we used Popoolation2 v1.201 (Kofler et al., 2011b) and breseq v0.35.0 (Deatherage and Barrick, 2014). BAM files from RGAPepPipe (see above) were filtered for quality and used with Samtools mpileup (Li et al., 2009) to tabulate mutation frequencies across sequenced timepoints. The mpileup file was converted to a sync file using Popoolation2 and finally converted the sync file to a format suitable for downstream analysis using Python (code available at https://github.com/myoungblom/mtb_ExpEvo, Smith, 2022 copy archived at swh:1:rev:5ed59742dc9219e32924c7450c77dc03b3663232). Variants were filtered for quality by minimum coverage of 20×, minimum alternate allele count of 5, and a minimum allele frequency of 5%. We also removed variants in repetitive families of genes (PE, PPE, and PE-PGRS) from analysis. Sequence data were also assembled and variants called and annotated using breseq v0.35.0 (Deatherage and Barrick, 2014) to confirm variants called using Popoolation2. Breseq was run in population mode for each sequenced timepoint, with all default parameters except one: ‘polymorphism-minimum-total-coverage-each-strand’ was set to 4. Then the mutational frequencies of each timepoint were compared using gdtools (Deatherage and Barrick, 2014) compare run with default parameters. To identify mutations that had significant frequency changes throughout the experiment, we looked for variants that were at a low starting frequency and rose to fixation (>95% frequency) or conversely, started at intermediate frequency and disappeared from the population (<5% frequency).
Structural variant detection
To identify potential insertions, deletions, and duplications of interest in our genomic sequence data, we called structural variants using Pilon v1.16 as a part of RGAPepPipe. We removed all insertions, deletions, and duplications that were located entirely in regions annotated as repetitive families of genes (PE, PPE, and PE-PGRS). We also removed variants that were identified in ancestral strains and/or that did not remain fixed in subsequent passages. Additionally, we manually curated potential variants by visually inspecting the alignments in Integrative Genomics Viewer (IGV) (Robinson et al., 2011; Thorvaldsdottir et al., 2012) and removed variants that appeared to be mis-mappings or in regions of poor coverage. Sliding window coverage plots to visualize duplications were made using Samtools bedcov (Li et al., 2009) using in-house scripts (code available at https://github.com/myoungblom/mtb_ExpEvo, Smith, 2022 copy archived at swh:1:rev:5ed59742dc9219e32924c7450c77dc03b3663232). Relative coverage was calculated by dividing each window coverage by the average coverage across the assembly – as given by BamQC (Okonechnikov et al., 2016) – and plotted in R.
GO term enrichment analysis
To identify possible functional enrichments in the genes within the large duplication that evolved in MT31 and MT55 pellicles (Figure 5), we performed a GO term enrichment analysis. GO terms for all M. tb genes were downloaded from Quick GO (https://www.ebi.ac.uk/QuickGO/). We used the R package topGO (Alexa and Rahnenfuhrer, 2022) to perform an enrichment analysis of the genes present in one or both duplications (a total of 172 genes). We calculated significance using a Fisher’s exact test and filtering for p<0.01 (Supplementary file 3). We performed the same enrichment analysis on 100 randomly selected, contiguous sets of 172 genes to test the greater significance of our results (Figure 8—figure supplement 2).
Genotype plots
To better understand the structure of the population over passaging, we created genotypes by clustering variants with similar trajectories throughout the experiment. Output from Popoolation2, as described above, was filtered for based on ≥30% frequency change between the final passage and ancestral populations, as well as filtering out all mutations within 1000 bp of each other (we identified that these mutations are often the result of repetitive regions and/or poor mapping). Lolipop (cdeitrick and pepepdodiu, 2020) was used with default parameters to cluster mutations into genotypes and create plots.
Population genetics statistics
Population genetics estimates (Tajima’s D, pi, and theta) were calculated as previously described (O’Neill et al., 2015). Briefly, we used Popoolation (Kofler et al., 2011a) to estimate these parameters in sliding windows across the genome. To reduce biases caused by variable coverage, we randomly sub-sampled read data without replacement to uniform coverage of 50×; we repeated this process nine times to further reduce biases. Genome-wide averages were calculated by averaging all windows across the genome, across all replicate sub-sampled data. Scripts available at https://github.com/myoungblom/mtb_ExpEvo, (Smith, 2022 copy archived at swh:1:rev:5ed59742dc9219e32924c7450c77dc03b3663232).
Network analysis
GOI were defined as coding genes containing or just downstream of the fixed mutations, we identified in populations passaged as pellicles (Figure 5). Network analysis of GOI was performed by accessing the TFOE data available in the MTB Network Portal (http://networks.systemsbiology.net/mtb/content/TFOE-Searchable-Data-File). TFs identified as significantly contributing to regulation of more than one of our GOIs were included in Figure 9. We also used this same dataset to identify GOIs which are themselves regulators and affect expression of other GOIs.
Fixed SNPs in global datasets
We searched for the presence of mutations identified in our study in publicly available sequencing data using a searchable Compact bit-sliced signature (COBS) index of bacterial genomes curated from the European Nucleotide Archive (Blackwell et al., 2021). We used the Python interface to search the COBS index (Bingmann et al., 2019). Our query sequences included 50 bp in either direction of the SNP of interest, with a k-mer matching threshold of 1. To determine the frequency of isolates in the COBS index with each SNP of interest, we needed to determine the number of M. tb genomes in the database: we searched the database using the full H37Rv 16S rRNA sequence (accession: NR_102810.2) which returned 39,941 results – this total number of M. tb isolates was used to calculate frequency of our SNPs of interest (Figure 5).
RNA isolation
Frozen cryovials of ancestral populations (MT540-0, MT49-0) and evolved populations (MT540-4, MT49-12) were thawed, and 250 µL of each was inoculated into 25 mL 7H9OTG and incubated at 37°C with shaking and grown to an OD600 of ~1. 250 µL of culture was used to inoculate 25 mL Sauton’s for biofilm growth. The remaining culture was pelleted, resuspended in 3 mL RNAprotect Bacteria Reagent (Qiagen), divided into 3×1 mL aliquots (three technical replicates), and frozen at –80°C until RNA extraction. Biofilm cultures were grown as previously described, and pellicles were harvested at 5 weeks. Pellicles were broken up and suspended in 3 mL RNAprotect, divided into 3×1 mL aliquots (three technical replicates), and frozen at –80°C until RNA extraction. RNA was extracted from each 1 mL aliquot (three technical replicates per sample), using the illustra RNAspin Mini RNA Isolation Kit (GE Healthcare) following the ‘Protocol for total RNA purification from up to 109 bacterial cells,’ which includes a 15 min room temperature incubation with RNase-free DNase I. RNA integrity was assessed by bleach gel electrophoresis (Aranda et al., 2012; ) and quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc).
Real-time quantitative qPCR
Approximately 100 ng of each RNA sample was used for cDNA synthesis with random hexamer primers using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) and included a 5 min incubation at 65°C and a reaction temperature of 44°C for GC-rich templates. Real-time quantitative PCR was performed in EU Fast Cycler PCR 96-well plates (BIOplastics) using the StepOnePlus Real-Time PCR System (v. 2.3, Applied Biosystems). We used sets of primers previously published and verified (Akhtar et al., 2009) to measure the expression of lpdA normalized to the expression of the endogenous control, sigA. All real-time PCR assays were run in a total reaction volume of 20 µL comprised 2× Fast SYBR Green Master Mix (Applied Biosystems), 200 nM of both forward and reverse primers (Integrated DNA Technologies, Coralville, IA, USA), and 3 µL of cDNA. RT-qPCR cycling parameters were set as follows: an initial AmpliTaq Fast DNA Polymerase, UP Activation step of 20 s at 95°C, followed by 40 cycles of 3 s at 95°C and 30 s at 60°C. Each reaction was repeated three times with three independent cDNA samples. Negative controls consisting of no-template reaction mixtures were run with all reactions. Melting-curve analysis was carried out to confirm the specificity of the amplified product. After baseline correction and determination of the threshold settings, the Factor-qPCR ratio method (Ruijter et al., 2015) was used to remove between-run variation due to the experiment including multiple plates. The factor corrected ∆CT values of the three technical replicates (MT540-EB has only 2 replicates) were then averaged, and the ∆∆CTs were calculated by subtracting a reference value (MT540-AP) from each sample. The expression fold change was calculated using the 2−∆∆CT method (Livak and Schmittgen, 2001), as PCR efficiencies had previously been found to be similar. Low and high fold changes were calculated by adding or subtracting the SD from the fold change. The results are expressed as log2 of the fold changes. Kruskal–Wallis test was used to determine the presence of significant log2 fold differences in the dataset (p-value=1.89e-04). Then, a Wilcoxon test was performed to compare log2 fold changes between sample types (AP, AB, EP, and EB) to identify significant differences between pairs. A p-value <0.05 was considered significant.
lpdA overexpression mutant
The lpdA gene was overexpressed in M. tb H37Rv at the L5 site using the integrative, constitutive overexpression plasmid pCT94 provided by Sarah Fortune’s lab (Harvard T.H. Chan School of Public Health). lpdA was amplified and modified to include NdeI and HindIII digestion sites using forward primer CCGCATGCTTAATTAAGAAGGAGATATACA-CCGCCGTCTAAGCTTTCCCCC and reverse primer GACCTCTAGGGTCCCCAATTAATTAGCTAA-TAGCCTGGGCGCTCGCGATAA. After amplification with Platinum SuperFi Green PCR Master Mix (Invitrogen, Waltham, MA), the PCR product was digested with NdeI and HindIII (NEB, Ipswich, MA). The digested product was then ligated with a NdeI and HindIII cut pCT94 using Gibson assembly (NEB, Ipswich, MA). The assembly product (CT94-lpdA) was then heat-shock transformed into DH5α Escherichia coli cells and selected for on LB plates with 50 μg/mL kanamycin overnight. Plasmid integration was verified via NdeI and HindIII digestion. CT94-lpdA and an empty vector control (CT94) were electroporated into electrocompetent M. tb H37Rv, and transformants were recovered in 2 mL of fresh 7H9 for 24–48 hr at 37°C, and then struck on 7H10 plates containing 25 μg/mL kanamycin. Both control and lpdA overexpression strains were grown as pellicles in triplicate and wet weight measurements taken as described above.
Acknowledgements
We would like to thank the Fortune lab with their assistance in the design of our overexpression experiments. We would like to acknowledge Soleil Young for her help phenotyping pellicles. We also thank the University of Wisconsin Biotechnology Center and the Microbial Genome Sequencing Center for providing sequencing services.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Caitlin S Pepperell, Email: cspepper@medicine.wisc.edu.
Dominique Soldati-Favre, University of Geneva, Switzerland.
Dominique Soldati-Favre, University of Geneva, Switzerland.
Funding Information
This paper was supported by the following grants:
National Institute of Allergy and Infectious Diseases R01AI113287 to Caitlin S Pepperell.
National Science Foundation DGE-1747503 to Madison A Youngblom.
National Institute of Allergy and Infectious Diseases T32-AI55391 to Lindsey L Bohr.
National Science Foundation DGE-1256259 to Tatum D Mortimer, Mary B O'Neill.
National Institute of Allergy and Infectious Diseases T32-GM07215 to Tatum D Mortimer.
Additional information
Competing interests
The authors declare no conflict of interes.
No competing interests declared.
Author contributions
Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review and editing.
Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review and editing.
Investigation, Visualization, Writing – review and editing.
Formal analysis, Writing – review and editing.
Investigation, Writing – review and editing.
Investigation, Writing – review and editing.
Investigation, Writing – review and editing.
Formal analysis, Writing – review and editing.
Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review and editing.
Ethics
Archived strains of Mycobacterium tuberculosis originally isolated from clinical specimens were analyzed in this study. Research on these strains was reviewed and approved by the University of Wisconsin Madison Institutional Review Board.
Additional files
Grouped by mutations within the same gene and sorted by genes that acquired the most independent SNPs across replicates and strains.
Of note is enrichment of nucleotide biosynthesis/metabolic processes.
‘Ref’ and ‘alt’ refer to the alleles present in the reference strain (H37Rv) and the clinical isolate, respectively. Mutations without an annotation exist in intergenic regions.
Data availability
Raw sequence data for ancestral and evolved isolates has been submitted to NCBI under the project accession PRJNA720149.
The following dataset was generated:
Smith TM, Youngblom MA. 2022. Experimental Evolution of Mycobacterium tuberculosis biofilms. NCBI BioProject. PRJNA720149
References
- Ackart DF, Hascall-Dove L, Caceres SM, Kirk NM, Podell BK, Melander C, Orme IM, Leid JG, Nick JA, Basaraba RJ. Expression of antimicrobial drug tolerance by attached communities of Mycobacterium tuberculosis. Pathogens and Disease. 2014;70:359–369. doi: 10.1111/2049-632X.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar P, Srivastava S, Srivastava A, Srivastava M, Srivastava BS, Srivastava R. Rv3303c of Mycobacterium tuberculosis protects tubercle bacilli against oxidative stress in vivo and contributes to virulence in mice. Microbes and Infection. 2006;8:2855–2862. doi: 10.1016/j.micinf.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Akhtar P, Singh S, Bifani P, Kaur S, Srivastava BS, Srivastava R. Variable-number tandem repeat 3690 polymorphism in Indian clinical isolates of Mycobacterium tuberculosis and its influence on transcription. Journal of Medical Microbiology. 2009;58:798–805. doi: 10.1099/jmm.0.002550-0. [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J. topGO. 3.15Bioconductor. 2022 https://bioconductor.org/packages/release/bioc/html/topGO.html
- Ali F, Seshasayee ASN. Dynamics of genetic variation in transcription factors and its implications for the evolution of regulatory networks in Bacteria. Nucleic Acids Research. 2020;48:4100–4114. doi: 10.1093/nar/gkaa162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. Babraham Bioinformatics; 2018. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Aranda PS, LaJoie DM, Jorcyk CL. Bleach gel: A simple agarose gel for analyzing RNA quality. Electrophoresis. 2012;33:366–369. doi: 10.1002/elps.201100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnvig KB, Comas I, Thomson NR, Houghton J, Boshoff HI, Croucher NJ, Rose G, Perkins TT, Parkhill J, Dougan G, Young DB, Bishai WR. Sequence-Based Analysis Uncovers an Abundance of Non-Coding RNA in the Total Transcriptome of Mycobacterium tuberculosis. PLOS Pathogens. 2011;7:e1002342. doi: 10.1371/journal.ppat.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon J, Alderwick LJ, Allnutt JA, Gabasova E, Watson R, Hatch KA, Clark SO, Jeeves RE, Marriott A, Rayner E, Tolley H, Pearson G, Hall G, Besra GS, Wernisch L, Williams A, Marsh PD, Tyagi AK. Non-Replicating Mycobacterium tuberculosis Elicits a Reduced Infectivity Profile with Corresponding Modifications to the Cell Wall and Extracellular Matrix. PLOS ONE. 2014;9:e87329. doi: 10.1371/journal.pone.0087329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. Inexpensive Multiplexed Library Preparation for Megabase-Sized Genomes. bioRxiv. 2015 doi: 10.1101/013771. [DOI] [PMC free article] [PubMed]
- Bharti S, Maurya RK, Venugopal U, Singh R, Akhtar M, Krishnan MY. Rv1717 Is a Cell Wall - Associated β-Galactosidase of Mycobacterium tuberculosis That Is Involved in Biofilm Dispersion. Frontiers in Microbiology. 2021;11:611122. doi: 10.3389/fmicb.2020.611122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingmann T, Bradley P, Gauger F, Iqbal Z. COBS: A Compact Bit-Sliced Signature Index. String Processing and Information Retrieval, Lecture Notes in Computer Science. 2019;1:285–303. doi: 10.1007/978-3-030-32686-9_21. [DOI] [Google Scholar]
- Blackwell GA, Hunt M, Malone KM, Lima L, Horesh G, Alako BTF, Thomson NR, Iqbal Z. Exploring bacterial diversity via a curated and searchable snapshot of archived DNA sequences. PLOS Biology. 2021;19:e3001421. doi: 10.1371/journal.pbio.3001421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad Institute . Broad Institute; 2022. http://broadinstitute.github.io/picard/ [Google Scholar]
- Brynildsrud OB, Pepperell CS, Suffys P, Grandjean L, Monteserin J, Debech N, Bohlin J, Alfsnes K, Pettersson JO-H, Kirkeleite I, Fandinho F, da Silva MA, Perdigao J, Portugal I, Viveiros M, Clark T, Caws M, Dunstan S, Thai PVK, Lopez B, Ritacco V, Kitchen A, Brown TS, van Soolingen D, O’Neill MB, Holt KE, Feil EJ, Mathema B, Balloux F, Eldholm V. Global expansion of Mycobacterium tuberculosis lineage 4 shaped by colonial migration and local adaptation. Science Advances. 2018;4:eaat5869. doi: 10.1126/sciadv.aat5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canetti G. Title The Tubercle Bacillus in the Pulmonary Lesion of Man. New York: Springer; 1955. [Google Scholar]
- Canetti G. THE TUBERCLE BACILLUS IN THE PULMONARY LESION OF MAN. The American Journal of the Medical Sciences. 1956;231:480. doi: 10.1097/00000441-195604000-00012. [DOI] [Google Scholar]
- Carey AF, Rock JM, Krieger IV, Chase MR, Fernandez-Suarez M, Gagneux S, Sacchettini JC, Ioerger TR, Fortune SM. TnSeq of Mycobacterium tuberculosis clinical isolates reveals strain-specific antibiotic liabilities. PLOS Pathogens. 2018;14:e1006939. doi: 10.1371/journal.ppat.1006939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro RAD, Ross A, Kamwela L, Reinhard M, Loiseau C, Feldmann J, Borrell S, Trauner A, Gagneux S. The Genetic Background Modulates the Evolution of Fluoroquinolone-Resistance in Mycobacterium tuberculosis. Molecular Biology and Evolution. 2020;37:195–207. doi: 10.1093/molbev/msz214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- cdeitrick, pepepdodiu Lolipop. 5b87b00GitHub. 2020 https://github.com/cdeitrick/Lolipop
- Chakraborty P, Bajeli S, Kaushal D, Radotra BD, Kumar A. Biofilm formation in the lung contributes to virulence and drug tolerance of Mycobacterium tuberculosis. Nature Communications. 2021;12:1606. doi: 10.1038/s41467-021-21748-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan R, Ravi J, Datta P, Chen T, Schnappinger D, Bassler KE, Balázsi G, Gennaro ML. Reconstruction and topological characterization of the sigma factor regulatory network of Mycobacterium tuberculosis. Nature Communications. 2016;7:11062. doi: 10.1038/ncomms11062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xie J. Role and regulation of bacterial LuxR-like regulators. Journal of Cellular Biochemistry. 2011;112:2694–2702. doi: 10.1002/jcb.23219. [DOI] [PubMed] [Google Scholar]
- Chiner-Oms Á, Berney M, Boinett C, González-Candelas F, Young DB, Gagneux S, Jacobs WR, Parkhill J, Cortes T, Comas I. Genome-wide mutational biases fuel transcriptional diversity in the Mycobacterium tuberculosis complex. Nature Communications. 2019;10:3994. doi: 10.1038/s41467-019-11948-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, Lewis NE, Landick R, Palsson BØ. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. PNAS. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial Biofilms: A Common Cause of Persistent Infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Deatherage DE, Barrick JE. Identification of mutations in laboratory evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. Clifton NJ. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb DK, Dahiya P, Srivastava KK, Srivastava R, Srivastava BS. Selective identification of new therapeutic targets of Mycobacterium tuberculosis by IVIAT approach. Tuberculosis. 2002;82:175–182. doi: 10.1054/tube.2002.0337. [DOI] [PubMed] [Google Scholar]
- DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, Ioerger TR, Stallings CL, Manoil C, Lampe D. Comprehensive Essentiality Analysis of the Mycobacterium tuberculosis Genome via Saturating Transposon Mutagenesis. MBio. 2017;8:e16. doi: 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P., Kolly GS, Leon-Solis L, Fallow A, Reed MB. Massive gene duplication event among clinical isolates of the Mycobacterium tuberculosis W/Beijing family. Journal of Bacteriology. 2010;192:4562–4570. doi: 10.1128/JB.00536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P, Rog A, Moolji J, Radomski N, Fallow A, Leon-Solis L, Bowes J, Behr MA, Reed MB. Origins of a 350-kilobase genomic duplication in Mycobacterium tuberculosis and its impact on virulence. Infection and Immunity. 2014;82:2902–2912. doi: 10.1128/IAI.01791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P., Zou J, Averback A, Syed N, Curtis D, Donato S, Reed MB. Unique Regulation of the DosR Regulon in the Beijing Lineage of Mycobacterium tuberculosis. Journal of Bacteriology. 2017;199:e00696-16. doi: 10.1128/JB.00696-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos RJ, Davis BD. FACTORS AFFECTING THE GROWTH OF TUBERCLE BACILLI IN LIQUID MEDIA. The Journal of Experimental Medicine. 1946;83:409–423. doi: 10.1084/jem.83.5.409. [DOI] [PubMed] [Google Scholar]
- Ektefaie Y, Dixit A, Freschi L, Farhat MR. Globally diverse Mycobacterium tuberculosis resistance acquisition: a retrospective geographical and temporal analysis of whole genome sequences. The Lancet. Microbe. 2021;2:e96–e104. doi: 10.1016/s2666-5247(20)30195-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldholm V, Balloux F. Antimicrobial Resistance in Mycobacterium tuberculosis: The Odd One Out. Trends in Microbiology. 2016;24:637–648. doi: 10.1016/j.tim.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Farhat MR, Freschi L, Calderon R, Ioerger T, Snyder M, Meehan CJ, de Jong B, Rigouts L, Sloutsky A, Kaur D, Sunyaev S, van Soolingen D, Shendure J, Sacchettini J, Murray M. GWAS for quantitative resistance phenotypes in Mycobacterium tuberculosis reveals resistance genes and regulatory regions. Nature Communications. 2019;10:2128. doi: 10.1038/s41467-019-10110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Kryazhimskiy S, Lang GI. Detecting genetic interactions using parallel evolution in experimental populations. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2019;374:20180237. doi: 10.1098/rstb.2018.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, Gomes A, Rustad T, Dolganov G, Glotova I, Abeel T, Mahwinney C, Kennedy AD, Allard R, Brabant W, Krueger A, Jaini S, Honda B, Yu WH, Hickey MJ, Zucker J, Garay C, Weiner B, Sisk P, Stolte C, Winkler JK, Van de Peer Y, Iazzetti P, Camacho D, Dreyfuss J, Liu Y, Dorhoi A, Mollenkopf HJ, Drogaris P, Lamontagne J, Zhou Y, Piquenot J, Park ST, Raman S, Kaufmann SHE, Mohney RP, Chelsky D, Moody DB, Sherman DR, Schoolnik GK. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013;499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrick ER, Barbier T, Chase MR, Xu R, François J, Lin VH, Szucs MJ, Rock JM, Ahmad R, Tjaden B, Livny J, Fortune SM. Small RNA profiling in Mycobacterium tuberculosis identifies MrsI as necessary for an anticipatory iron sparing response. PNAS. 2018;115:6464–6469. doi: 10.1073/pnas.1718003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Tuberculosis Report Global Tuberculosis programme. 2019. [November 3, 2021]. https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-report-2019
- Gomez-Gonzalez PJ, Andreu N, Phelan JE, de Sessions PF, Glynn JR, Crampin AC, Campino S, Butcher PD, Hibberd ML, Clark TG. An integrated whole genome analysis of Mycobacterium tuberculosis reveals insights into relationship between its genome, transcriptome and methylome. Scientific Reports. 2019;9:5204. doi: 10.1038/s41598-019-41692-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde SR. Computational Identification of the Proteins Associated With Quorum Sensing and Biofilm Formation in Mycobacterium tuberculosis. Frontiers in Microbiology. 2019;10:3011. doi: 10.3389/fmicb.2019.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. Stable association between strains of Mycobacterium tuberculosis and their human host populations. PNAS. 2004;101:4871–4876. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Coates ARM. Mycobacterium tuberculosis acg gene is required for growth and virulence in vivo. PLOS ONE. 2011;6:e20958. doi: 10.1371/journal.pone.0020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Systematic Biology. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Kerns PW, Ackhart DF, Basaraba RJ, Leid JG, Shirtliff ME. Mycobacterium tuberculosis pellicles express unique proteins recognized by the host humoral response. Pathogens and Disease. 2014;70:347–358. doi: 10.1111/2049-632X.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. Negative epistasis between beneficial mutations in an evolving bacterial population. Science (New York, N.Y.) 2011;332:1193–1196. doi: 10.1126/science.1203801. [DOI] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nature Reviews. Genetics. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Kobayashi K. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. Journal of Bacteriology. 2007;189:4920–4931. doi: 10.1128/JB.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Orozco-terWengel P, De Maio N, Pandey RV, Nolte V, Futschik A, Kosiol C, Schlötterer C. PoPoolation: A toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLOS ONE. 2011a;6:e15925. doi: 10.1371/journal.pone.0015925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Pandey RV, Schlötterer C. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq. Bioinformatics (Oxford, England) 2011b;27:3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F. Babraham Bioinformatics; 2019. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ [Google Scholar]
- Kulka K, Hatfull G, Ojha AK. Growth of Mycobacterium tuberculosis biofilms. Journal of Visualized Experiments. 2012;1:3820. doi: 10.3791/3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. [DOI]
- Li X, Lv X, Lin Y, Zhen J, Ruan C, Duan W, Li Y, Xie J. Role of two‐component regulatory systems in intracellular survival of Mycobacterium tuberculosis. Journal of Cellular Biochemistry. 2019;120:12197–12207. doi: 10.1002/jcb.28792. [DOI] [PubMed] [Google Scholar]
- Lipworth S, Jajou R, de Neeling A, Bradley P, van der Hoek W, Maphalala G, Bonnet M, Sanchez-Padilla E, Diel R, Niemann S, Iqbal Z, Smith G, Peto T, Crook D, Walker T, van Soolingen D. SNP-IT Tool for Identifying Subspecies and Associated Lineages of Mycobacterium tuberculosis Complex. Emerging Infectious Diseases. 2019;25:482–488. doi: 10.3201/eid2503.180894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Ma A, Wei L, Pang Y, Wu B, Luo T, Zhou Y, Zheng HX, Jiang Q, Gan M, Zuo T, Liu M, Yang C, Jin L, Comas I, Gagneux S, Zhao Y, Pepperell CS, Gao Q. China’s tuberculosis epidemic stems from historical expansion of four strains of Mycobacterium tuberculosis. Nature Ecology & Evolution. 2018;2:1982–1992. doi: 10.1038/s41559-018-0680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science (New York, N.Y.) 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Maksymiuk C, Ioerger T, Balakrishnan A, Bryk R, Rhee K, Sacchettini J, Nathan C. Comparison of transposon and deletion mutants in Mycobacterium tuberculosis: The case of rv1248c, encoding 2-hydroxy-3-oxoadipate synthase. Tuberculosis (Edinburgh, Scotland) 2015;95:689–694. doi: 10.1016/j.tube.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson AL, Cohen KA, Abeel T, Desjardins CA, Armstrong DT, Barry CE, Brand J, Chapman SB, Cho SN, Gabrielian A, Gomez J, Jodals AM, Joloba M, Jureen P, Lee JS, Malinga L, Maiga M, Nordenberg D, Noroc E, Romancenco E, Salazar A, Ssengooba W, Velayati AA, Winglee K, Zalutskaya A, Via LE, Cassell GH, Dorman SE, Ellner J, Farnia P, Galagan JE, Rosenthal A, Crudu V, Homorodean D, Hsueh PR, Narayanan S, Pym AS, Skrahina A, Swaminathan S, Van der Walt M, Alland D, Bishai WR, Cohen T, Hoffner S, Birren BW, Earl AM, TBResist Global Genome Consortium Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nature Genetics. 2017;49:395–402. doi: 10.1038/ng.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlin SJ, Conkle-Gutierrez D, Kim C, Mitchell SN, Morrissey C, Weinrick BC, Jacobs WR, Ramirez-Busby SM, Hoffner SE, Valafar F. Drivers and sites of diversity in the DNA adenine methylomes of 93 Mycobacterium tuberculosis complex clinical isolates. eLife. 2020;9:e58542. doi: 10.7554/eLife.58542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer TD, Weber AM, Pepperell CS. Signatures of Selection at Drug Resistance Loci in Mycobacterium tuberculosis. MSystems. 2018;3:e00108-17. doi: 10.1128/mSystems.00108-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland CV, Shockey AC, Aung HL, Cursons RT, O’Toole RF, Gautam SS, Brites D, Gagneux S, Roberts SA, Karalus N, Cook GM, Pepperell CS, Arcus VL. Dispersal of Mycobacterium tuberculosis Driven by Historical European Trade in the South Pacific. Frontiers in Microbiology. 2019;10:2778. doi: 10.3389/fmicb.2019.02778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyka W. STUDIES ON MYCOBACTERIUM TUBERCULOSIS IN LESIONS OF THE HUMAN LUNG: A NEW METHOD OF STAINING TUBERCLE BACILLI IN TISSUE SECTIONS. The American Review of Respiratory Disease. 1963;88:670–679. doi: 10.1164/arrd.1963.88.5.670. [DOI] [PubMed] [Google Scholar]
- Nyka W. Method for staining both acid-fast and chromophobic tubercle bacilli with carbolfuschsin. Journal of Bacteriology. 1967;93:1458–1460. doi: 10.1128/jb.93.4.1458-1460.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyka W, O’Neill EF. A NEW APPROACH TO THE STUDY OF NON-ACID-FAST MYCOBACTERIA. Annals of the New York Academy of Sciences. 1970;174:862–871. doi: 10.1111/j.1749-6632.1970.tb45605.x. [DOI] [PubMed] [Google Scholar]
- Nyka W. The chromophobic tubercle bacilli and the problem of endogenous reactivation of tuberculosis. Materia Medica Polona. Polish Journal of Medicine and Pharmacy. 1977;9:175–185. [PubMed] [Google Scholar]
- Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs WR, Hatfull GF. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Molecular Microbiology. 2008;69:164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics (Oxford, England) 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Øyås O, Borrell S, Trauner A, Zimmermann M, Feldmann J, Liphardt T, Gagneux S, Stelling J, Sauer U, Zampieri M. Model-based integration of genomics and metabolomics reveals SNP functionality in Mycobacterium tuberculosis. PNAS. 2020;117:8494–8502. doi: 10.1073/pnas.1915551117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill MB, Mortimer TD, Pepperell CS. Diversity of Mycobacterium tuberculosis across Evolutionary Scales. PLOS Pathogens. 2015;11:e1005257. doi: 10.1371/journal.ppat.1005257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill MB, Shockey A, Zarley A, Aylward W, Eldholm V, Kitchen A, Pepperell CS. Lineage specific histories of Mycobacterium tuberculosis dispersal in Africa and Eurasia. Molecular Ecology. 2019;28:3241–3256. doi: 10.1111/mec.15120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, Harris SR. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microbial Genomics. 2016;2:e56. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JM, Layre E, Sweet L, Sherrid A, Moody DB, Ojha A, Sherman DR. The Polyketide Pks1 Contributes to Biofilm Formation in Mycobacterium tuberculosis. Journal of Bacteriology. 2012;194:715–721. doi: 10.1128/JB.06304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish T, Smith DA, Kendall S, Casali N, Bancroft GJ, Stoker NG. Deletion of Two-Component Regulatory Systems Increases the Virulence of Mycobacterium tuberculosis. Infection and Immunity. 2003;71:1134–1140. doi: 10.1128/IAI.71.3.1134-1140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvati Sai Arun PV, Miryala SK, Rana A, Kurukuti S, Akhter Y, Yellaboina S. System-wide coordinates of higher order functions in host-pathogen environment upon Mycobacterium tuberculosis infection. Scientific Reports. 2018;8:5079. doi: 10.1038/s41598-018-22884-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperell C, Hoeppner VH, Lipatov M, Wobeser W, Schoolnik GK, Feldman MW. Bacterial genetic signatures of human social phenomena among M. tuberculosis from an Aboriginal Canadian population. Molecular Biology and Evolution. 2010;27:427–440. doi: 10.1093/molbev/msp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperell CS, Granka JM, Alexander DC, Behr MA, Chui L, Gordon J, Guthrie JL, Jamieson FB, Langlois-Klassen D, Long R, Nguyen D, Wobeser W, Feldman MW. Dispersal of Mycobacterium tuberculosis via the Canadian fur trade. PNAS. 2011;108:6526–6531. doi: 10.1073/pnas.1016708108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperell CS, Casto AM, Kitchen A, Granka JM, Cornejo OE, Holmes EC, Holmes EC, Birren B, Galagan J, Feldman MW. The role of selection in shaping diversity of natural M. tuberculosis populations. PLOS Pathogens. 2013;9:e1003543. doi: 10.1371/journal.ppat.1003543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Lago L, Navarro Y, Herranz M, Bouza E, García-de-Viedma D. Differences in gene expression between clonal variants of Mycobacterium tuberculosis emerging as a result of microevolution. International Journal of Medical Microbiology. 2013;303:674–677. doi: 10.1016/j.ijmm.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Phelan J, de Sessions PF, Tientcheu L, Perdigao J, Machado D, Hasan R, Hasan Z, Bergval IL, Anthony R, McNerney R, Antonio M, Portugal I, Viveiros M, Campino S, Hibberd ML, Clark TG. Methylation in Mycobacterium tuberculosis is lineage specific with associated mutations present globally. Scientific Reports. 2018;8:160. doi: 10.1038/s41598-017-18188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N, Crozat E, Lenski RE, Schneider D. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology. 2007;29:846–860. doi: 10.1002/bies.20629. [DOI] [PubMed] [Google Scholar]
- Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445:383–386. doi: 10.1038/nature05451. [DOI] [PubMed] [Google Scholar]
- Rastogi S, Singh AK, Pant G, Mitra K, Sashidhara KV, Krishnan MY. Down-regulation of PE11, a cell wall associated esterase, enhances the biofilm growth of Mycobacterium tuberculosis and reduces cell wall virulence lipid levels. Microbiology (Reading, England) 2017;163:52–61. doi: 10.1099/mic.0.000417. [DOI] [PubMed] [Google Scholar]
- Richards JP, Cai W, Zill NA, Zhang W, Ojha AK. Adaptation of Mycobacterium tuberculosis to Biofilm Growth Is Genetically Linked to Drug Tolerance. Antimicrobial Agents and Chemotherapy. 2019;63:e01213-19. doi: 10.1128/AAC.01213-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Verdugo A, Tenaillon O, Gaut BS. First-Step Mutations during Adaptation Restore the Expression of Hundreds of Genes. Molecular Biology and Evolution. 2016;33:25–39. doi: 10.1093/molbev/msv228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ruiz Villalba A, Hellemans J, Untergasser A, van den Hoff MJB. Removal of between-run variation in a multi-plate qPCR experiment. Biomolecular Detection and Quantification. 2015;5:10–14. doi: 10.1016/j.bdq.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad TR, Minch KJ, Ma S, Winkler JK, Hobbs S, Hickey M, Brabant W, Turkarslan S, Price ND, Baliga NS, Sherman DR. Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biology. 2014;15:502. doi: 10.1186/PREACCEPT-1701638048134699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandan D, Dao DN, Weinrick BC, Vilchèze C, Gurcha SS, Ojha A, Kremer L, Besra GS, Hatfull GF, Jacobs WR. Keto-mycolic acid-dependent pellicle formation confers tolerance to drug-sensitive Mycobacterium tuberculosis. MBio. 2013;4:e00213. doi: 10.1128/mBio.00222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxer G, Krepps MD, Merkley ED, Ansong C, Deatherage Kaiser BL, Valovska M-T, Ristic N, Yeh PT, Prakash VP, Leiser OP, Nakhleh L, Gibbons HS, Kreuzer HW, Shamoo Y. Mutations in global regulators lead to metabolic selection during adaptation to complex environments. PLOS Genetics. 2014;10:e1004872. doi: 10.1371/journal.pgen.1004872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma IM, Petchiappan A, Chatterji D. Quorum sensing and biofilm formation in mycobacteria: role of c-di-GMP and methods to study this second messenger. IUBMB Life. 2014;66:823–834. doi: 10.1002/iub.1339. [DOI] [PubMed] [Google Scholar]
- Shell SS, Prestwich EG, Baek SH, Shah RR, Sassetti CM, Dedon PC, Fortune SM. DNA methylation impacts gene expression and ensures hypoxic survival of Mycobacterium tuberculosis. PLOS Pathogens. 2013;9:e1003419. doi: 10.1371/journal.ppat.1003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva RF, Mendonça SCM, Carvalho LM, Reis AM, Gordo I, Trindade S, Dionisio F. Pervasive sign epistasis between conjugative plasmids and drug-resistance chromosomal mutations. PLOS Genetics. 2011;7:e1002181. doi: 10.1371/journal.pgen.1002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Sharma M, Chaudhry A, Sharma S. Rv2626c and Rv2032 activate TH1 response and downregulate regulatory T cells in peripheral blood mononuclear cells of tuberculosis patients. Comparative Immunology, Microbiology and Infectious Diseases. 2019;62:46–53. doi: 10.1016/j.cimid.2018.11.016. [DOI] [PubMed] [Google Scholar]
- Smith TM. mtb_ExpEvo. swh:1:rev:5ed59742dc9219e32924c7450c77dc03b3663232Software Heritage. 2022 https://archive.softwareheritage.org/swh:1:dir:c24b59e53d7c132602d07dfcf9712c4f2ba20145;origin=https://github.com/myoungblom/mtb_ExpEvo;visit=swh:1:snp:112a276f639a0936c57da74187b84c0b441d297d;anchor=swh:1:rev:5ed59742dc9219e32924c7450c77dc03b3663232
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe HA, Bayliss SC, Hurst LD, Feil EJ. Comparative Analyses of Selection Operating on Nontranslated Intergenic Regions of Diverse Bacterial Species. Genetics. 2017;206:363–376. doi: 10.1534/genetics.116.195784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics. 2012;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizzano B, Dallenga TK, Utpatel C, Behrends J, Homolka S, Kohl TA, Niemann S. Survival of hypoxia-induced dormancy is not a common feature of all strains of the Mycobacterium tuberculosis complex. Scientific Reports. 2021;11:2628. doi: 10.1038/s41598-021-81223-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trauner A, Liu Q, Via LE, Liu X, Ruan X, Liang L, Shi H, Chen Y, Wang Z, Liang R, Zhang W, Wei W, Gao J, Sun G, Brites D, England K, Zhang G, Gagneux S, Barry CE, Gao Q. The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy. Genome Biology. 2017;18:71. doi: 10.1186/s13059-017-1196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi A, Mavi PS, Bhatt D, Kumar A. Thiol reductive stress induces cellulose-anchored biofilm formation in Mycobacterium tuberculosis. Nature Communications. 2016;7:11392. doi: 10.1038/ncomms11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Sambandan D, Halder R, Wang J, Batt SM, Weinrick B, Ahmad I, Yang P, Zhang Y, Kim J, Hassani M, Huszar S, Trefzer C, Ma Z, Kaneko T, Mdluli KE, Franzblau S, Chatterjee AK, Johnsson K, Johnson K, Mikusova K, Besra GS, Fütterer K, Robbins SH, Barnes SW, Walker JR, Jacobs WR, Schultz PG. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. PNAS. 2013;110:E2510–E2517. doi: 10.1073/pnas.1309171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner B, Gomez J, Victor TC, Warren RM, Sloutsky A, Plikaytis BB, Posey JE, van Helden PD, Gey van Pittius NC, Koehrsen M, Sisk P, Stolte C, White J, Gagneux S, Birren B, Hung D, Murray M, Galagan J. Independent large scale duplications in multiple M. tuberculosis lineages overlapping the same genomic region. PLOS ONE. 2012;7:e26038. doi: 10.1371/journal.pone.0026038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Watson RA, Chao L. Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution; International Journal of Organic Evolution. 2005;59:1165–1174. [PubMed] [Google Scholar]
- Wolff KA, de la Peña AH, Nguyen HT, Pham TH, Amzel LM, Gabelli SB, Nguyen L. A redox regulatory system critical for mycobacterial survival in macrophages and biofilm development. PLOS Pathogens. 2015;11:e1004839. doi: 10.1371/journal.ppat.1004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Blair KM, Salama NR. Staying in Shape: the Impact of Cell Shape on Bacterial Survival in Diverse Environments. Microbiology and Molecular Biology Reviews. 2016;80:187–203. doi: 10.1128/MMBR.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Thomas J, Li Y, Vilchèze C, Derbyshire KM, Jacobs WR, Ojha AK. Defining a temporal order of genetic requirements for development of mycobacterial biofilms. Molecular Microbiology. 2017;105:794–809. doi: 10.1111/mmi.13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblom M. RGAPepPipe. 0122dcaGitHub. 2021 https://github.com/pepperell-lab/RGAPepPipe
- Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends in Ecology & Evolution. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- Zheng H, Lu L, Wang B, Pu S, Zhang X, Zhu G, Shi W, Zhang L, Wang H, Wang S, Zhao G, Zhang Y. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLOS ONE. 2008;3:e2375. doi: 10.1371/journal.pone.0002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhong J, Jia X, Liu G, Kang Y, Dong M, Zhang X, Li Q, Yue L, Li C, Fu J, Xiao J, Yan J, Zhang B, Lei M, Chen S, Lv L, Zhu B, Huang H, Chen F. Precision methylome characterization of Mycobacterium tuberculosis complex (MTBC) using PacBio single-molecule real-time (SMRT) technology. Nucleic Acids Research. 2016;44:730–743. doi: 10.1093/nar/gkv1498. [DOI] [PMC free article] [PubMed] [Google Scholar]