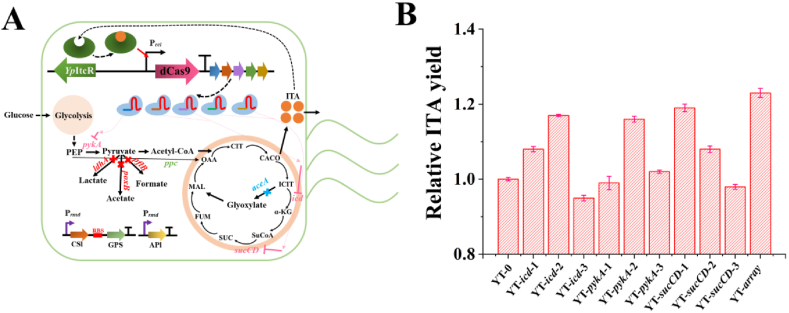
Fig. 4.
Biosynthesis of itaconic acid (ITA) for the dynamic regulation of metabolic flux by the CRISPRi-mediated self-inducible system (CiMS). (A) Construction of different sgRNAs targeted to essential genes to interfere with the metabolic pathway. CSl (CAD-SH3lig), GPS (GA-PDZ-SH3), and APl (ACN-PDZlig) represent the over-expression cascade reaction for ITA production. The genes marked with blue and red represent the genes knocked out in the genome of Δ4-Prmd-SAS-3. The genes marked with a pink color represent the target genes repressed by the CiMS. After accumulating sufficient ITA, the ITA-bound YpItcR could activate the expression of dCas9 and inhibit the expression of the genes in pink with the help of the sgRNA. (B) Relative ITA yield of the CiMS-regulated strains with different sgRNAs. Escherichia coli MG1655 Δ4-Prmd-SAS-3/sgRNA-0 was used as the control. The sgRNA-array contained five sgRNAs, sgRNA-icd-1, sgRNA-icd-2, sgRNA-pykA-1, sgRNA-sucCD-1, sgRNA-sucCD-2, the inhibition of which could significantly improve the production of ITA. Error bars show the standard deviation from three independent experiments.