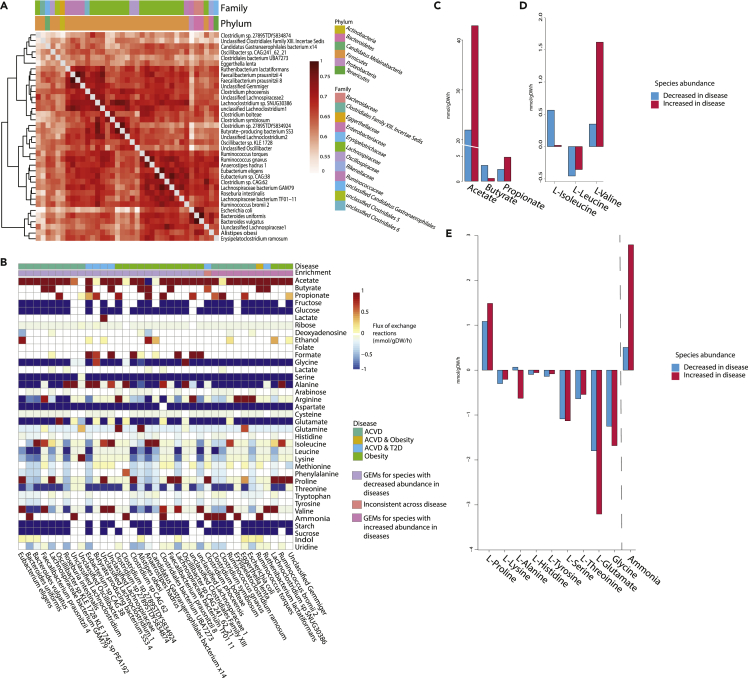
Figure 2.
An overview of the individual genome-scale models and their individual fluxes
(A) Jaccard index of the models, showing similarity between the different species based on reactions. Distances range 0-1 a score of one represents two models which are identical, a score of 0 represents two models which have no overlapping reactions.
(B) Heatmap showing the flux for different metabolites of the models constrained by a high fiber omnivore diet, with the objective function to optimize biomass, using COBRA Toolbox. For visualization, flux was not shown for greater or lesser than +/−1.
(C) The average flux from the models which were increased in disease (pink) compared to the average flux from the models decreased in disease (blue) for SCFAs.
(D) The average flux from the models which were increased in disease (pink) compared to the average flux from the models decreased in disease (blue) for BCAAs.
(E) The average flux from the models increased in disease (pink) compared to the average flux from the models decreased in disease (blue) for metabolites that show a difference between the average flux amounts. For C-E negative flux implies the metabolite is up taken by the model, positive flux implies the model secretes this metabolite.