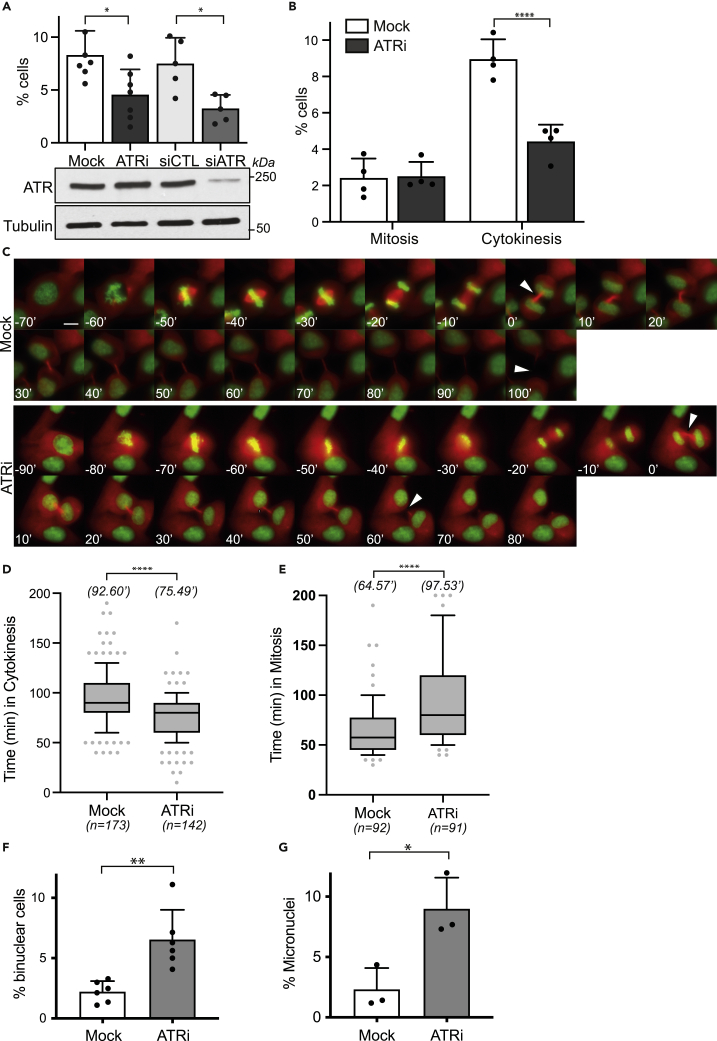
Figure 3.
ATR is a negative regulator of abscission required for genome stability
(A) Percentage of cytokinetic cells upon pharmacological inhibition (10 μM ETP-46464; 1 h) or genetic depletion of ATR. The Western blots indicate the expression levels of ATR and the α-tubulin loading control. At least, 200 cells were analyzed in each of a minimum of 5 independent experiments (∗p = 0.0214, ∗p = 0.0029; Student’s t-test).
(B) Percentage of mitotic and cytokinetic cells upon pharmacological inhibition of ATR (ETP-46464, 10 μM; 1h). At least, 400 cells were analyzed in each of 4 independent experiments (∗∗∗∗p = 0.0001, Student’s t-test).
(C) Selected frames of single cells from live cell microscopy using asynchronously growing HeLa cells stably expressing GFP-H2B and mCherry-α-tubulin. Cells were treated with DMSO (mock) or ATR inhibitor (ETP-46464, 10 μM) immediately prior to a 16 h time-lapse experiment. Mitosis was measured from visible chromosome condensation in prophase to separation of chromosomes in telophase. Abscission timing was determined by measuring time from a visible cytoplasmic canal separating daughter cells with decondensed chromatin to complete microtubule severance.
(D) Quantification of abscission timing illustrated in (C). The number of cells analyzed across 3 independent experiments is indicated below each column (∗∗∗∗p = 0.0001, Student’s t-test).
(E) Quantification of mitotic timing illustrated in (C). The number of cells analyzed across 3 independent experiments is indicated below each column (∗∗∗∗p = 0.0001, Student’s t-test).
(F) Quantification of binuclear cells in response to ATR inhibition (ETP-46464, 10 μM) across six independent experiments (∗∗p = 0.0024, Student’s t-test).
(G) Quantification of micronuclei in response to ATR inhibition (ETP-46464, 10 μM) across three independent experiments (∗p = 0.0210, Student’s t-test). Data are represented as mean ± SD of the individual replicates in panels A,B, F, and G indicate SD, boxplot of panels in E and F indicate the median and quartile ranges. HeLa cells were used in A–G. All scale bars are 10 μm. See also Figure S3.