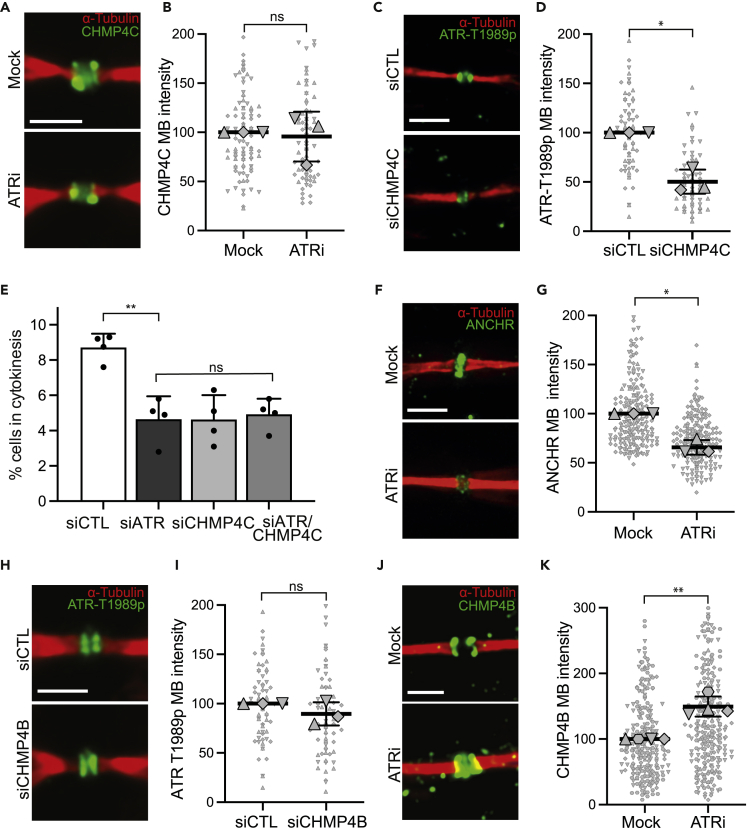
Figure 5.
ATR functions downstream of CHMP4C but upstream of CHMP4B and ANCHR in the regulation of abscission
(A) Localization of CHM4PC to the midbody of late cytokinetic cells upon ATR inhibition (ETP-46464, 10 μM; 1 h).
(B) Quantification of CHMP4C intensity at the midbody of late cytokinetic cells upon ATR inhibition (ETP-46464, 10 μM; 1 h),(ns, Student’s t-test).
(C) Localization of ATR-T1989p to late cytokinetic midbodies upon depletion of CHMP4C.
(D) Quantification of ATR-T1989p intensity at late cytokinetic midbodies upon depletion of CHMP4C. (∗p = 0.0195; Student’s t-test).
(E) Percentage of cells in cytokinesis upon depletion of ATR and/or CHMP4C. At least, 400 cells were analyzed in each of 4 independent experiments (∗∗p = 0.0017; Student’s t-test).
(F) Localization of ANCHR to late cytokinetic midbodies upon ATR inhibition (ETP-46464, 10 μM; 1 h).
(G) Quantification of ANCHR intensity at late cytokinetic midbodies upon ATR inhibition (ETP-46464, 10 μM; 1 h). (∗p = 0.0153; Student’s t-test).
(H) Localization of ATR-T1989p to late cytokinetic midbodies upon depletion of CHMP4B.
(I) Quantification of ATR-T1989p intensity at late cytokinetic midbodies upon depletion of CHMP4B. (ns; Student’s t-test).
(J) Localization of CHM4B to the midbody of late cytokinetic cells upon ATR inhibition (ETP-46464, 10 μM; 1 h).
(K) Quantification of CHM4B intensity at the midbody of late cytokinetic cells upon ATR inhibition (ETP-46464, 10 μM; 1 h). (∗∗p = 0.0077; Student’s t-test). All dot plots represent data from three independent biological replicates, each replicate is represented by a different symbol. All data have been normalized to the control sample. Data are represented as mean ± SD of the individual replicates. All scale bars are 10 μm, all enlarged image scale bars are 2 μm. HeLa cells were used in A–K. See also Figure S5.