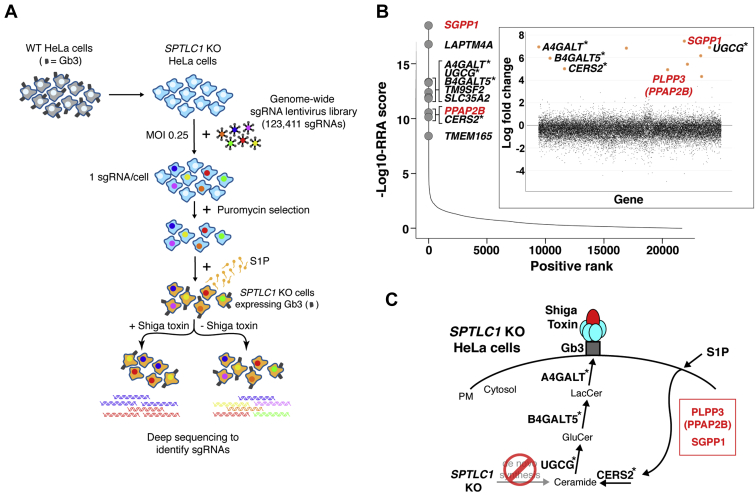
Fig. 2.
Genome-wide CRISPR/Cas9 KO screen identifies regulators of S1P cellular uptake and recycling into the sphingolipid synthesis pathway. A: Schematic of the genome-wide CRISPR/Cas9 KO screening strategy undertaken to identify genes required for S1P uptake and recycling. WT Cas9-expressing HeLa cells (gray); SPTLC1 KO Cas9-expressing HeLa cells (light blue). Cells were transduced (MOI of 0.25) with a lentivirus-based genome-wide CRISPR/Cas9 KO library and selected in puromycin to yield cells that each contained approximately one viral genome. Cells were grown in media containing S1P (orange) to induce Gb3 cell-surface expression and treated either with or without Shiga toxin. The genomic DNA of cells from each group was subjected to deep sequencing to identify the sgRNAs. B: Scatterplot showing the ranking of genes from MAGeCK analysis. X-axis indicates positive ranking of individual genes, and y-axis indicates -Log10 values of corresponding robust ranking aggregation (RRA) score. The 10 top-ranking genes are highlighted and labeled. Inset panel, scatterplot showing log-fold change for all genes. The genes for enzymes directly involved in Gb3 synthesis are marked with asterisks. The genes shown in red are involved in the S1P uptake and recycling pathway. C: Schematic showing sphingolipid synthesis pathway leading to the production of Gb3 in SPTLC1 KO HeLa cells. Top-ranking genes from the screen that are directly involved in Gb3 synthesis are marked with asterisks. PLPP3 (PPAP2B) and SGPP1, which are not involved in de novo synthesis of Gb3 (20, 21, 22, 23), are involved in the S1P uptake and recycling pathway. CRISPR, clustered regularly interspersed short palindromic repeats; Gb3, globotriaosylceramide; MAGeCK, Model-based Analysis of Genome-wide CRISPR-Cas9 KO; MOI, multiplicity of infection; PM, plasma membrane; S1P, sphingosine-1-phosphate; sgRNA, single guide RNA.