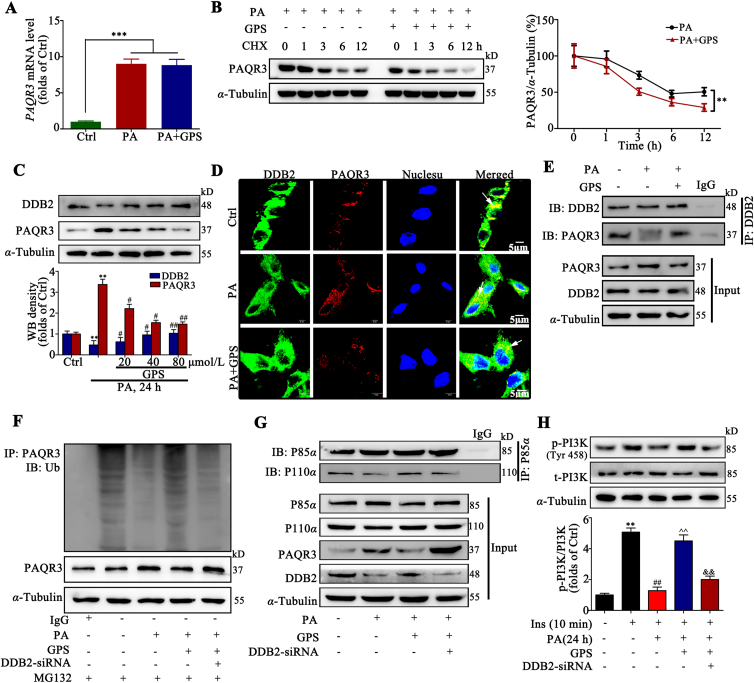
Figure 3.
GPS increased the ubiquitination of PAQR3 by promoting the interaction between DDB2 and PAQR3. (A) PAQR3 mRNA level in HepG2 cells (n = 3). ∗∗∗P < 0.001 vs. Ctrl. (B) The protein level of PAQR3 was determined by Western blot assay with HepG2 cells treated with PA and/or GPS in the presence of CHX (50 mg/mL) for 0, 1, 2, 4, 6, and 12 h (n = 3). ∗∗P < 0.001 vs. PA-treated group. (C) DDB2 and PAQR3 protein expression in HepG2 cells (n = 3). ∗∗P < 0.01 vs. Ctrl; #P < 0.05, ##P < 0.01 vs. PA-treated group. (D) Immunofluorescence staining presents the distribution and co-location of DDB2 and PAQR3 in HepG2 cells, scale bar = 20 μm. (E) A co-immunoprecipitation assay was performed to detect the interaction of DDB2 and PAQR3 in HepG2 cells. (F) Knockdown DDB2 decreased the polyubiquitination of PAQR3 induced by GPS in HepG2 cells. (G) The interaction of p85α and p110α when HepG2 cells were treated with PA and/or GPS, in the presence of PAQR3 siRNA. (H) Phosphorylation level of PI3K in HepG2 cells (n = 3). ∗∗P < 0.01 vs. Ctrl; ##P < 0.01 vs. insulin-treated group; ˆˆP < 0.01 vs. insulin + PA-treated group; &&P < 0.01 vs. insulin + PA + GPS-treated group. Protein was quantified and normalized to α-tubulin in the control group cells. The data are presented as mean ± SD, all experiments were performed at least three times with similar results.