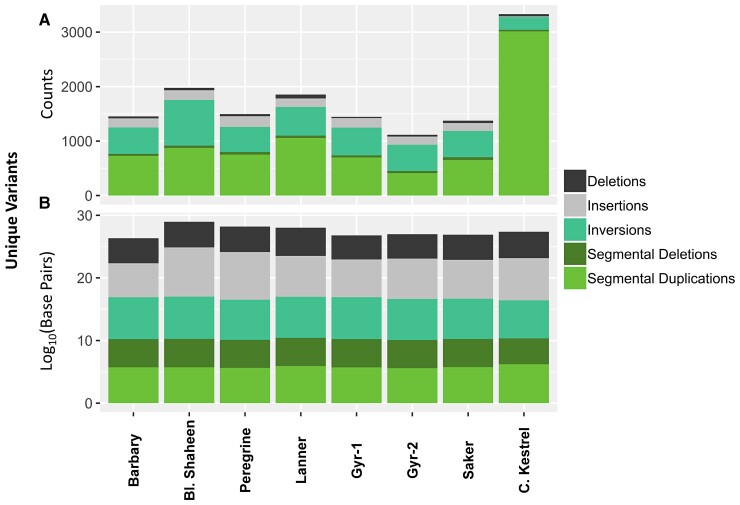
Fig. 1.
Unique structural variants within each genome based on MUMmer alignments by: (A) count; (B) log10(base pairs). Reported numbers are for diploid genomes and are color coded by type of structural variant. Structural variants are determined by alignment to the common kestrel outgroup. Segmental insertions and deletions are distinguished from insertions and deletions in that they represent the addition or disappearance of linked repetitive content on the same scaffold.