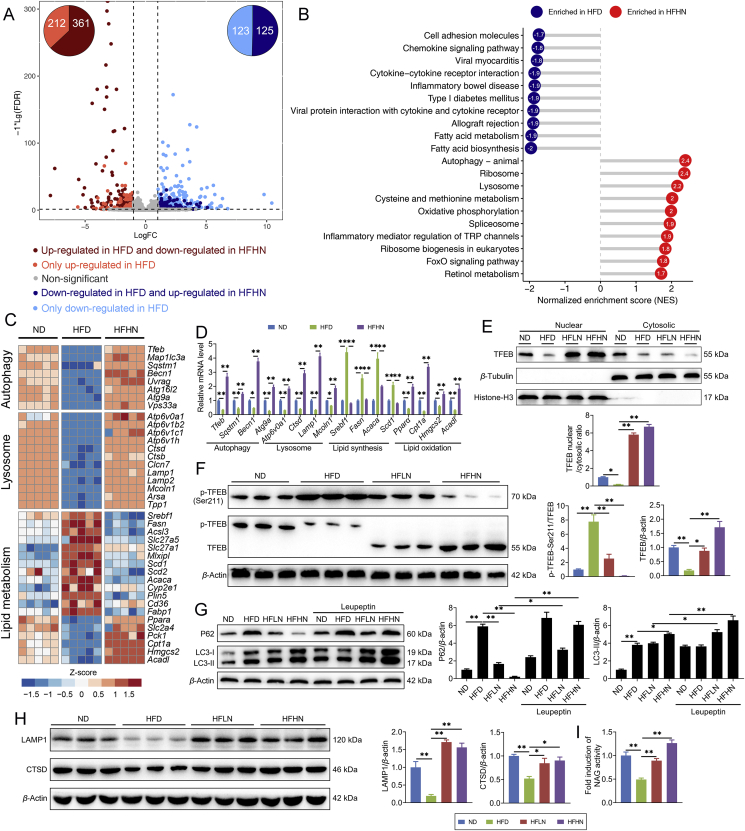
Figure 2.
Nuciferine activates hepatic autophagy–lysosomal pathway (ALP) in NAFLD mice. (A) The scatterplot showing fold change of all genes in the RNA-seq dataset, with differentially expressed genes shown with corresponding font color. The pie charts indicate the number of differentially expressed genes regulated by nuciferine and HFD feeding. (B) Gene set enrichment analysis showing the cellular pathways, which were upregulated by HFD but downregulated by nuciferine treatment. (C) Heatmap showing the expression profile of genes related to autophagy, lysosome and lipid metabolism in different groups. (D) Relative mRNA levels of the indicated genes were normalized to β-actin and Gapdh and expressed as fold change relative to ND group. (E) Representative immunoblotting of nuclear and cytosolic TFEB in the livers from different groups and quantification of TFEB nuclear/cytosolic ratio and expressed as fold change relative to ND group. (F) Representative immunoblotting of transcription factor EB (TFEB) and p-TFEB-Ser211 in the livers from different groups and quantification of p-TFEB-Ser211/TFEB, TFEB/β-actin and expressed as fold change relative to ND group. (G) Representative immunoblotting of sequestosome 1 (P62) and microtubule associated protein 1 light chain 3 (LC3) in the livers from different groups and quantification of P62/β-actin, LC3-II/β-actin and expressed as fold change relative to ND group. Mice were treated as described in Fig. 1 and sacrificed 8 h after intraperitoneal injection of leupeptin (40 mg/kg). (H) Representative immunoblotting of lysosomal associated membrane protein 1 (LAMP1) and cathepsin D (CTSD) in the livers from different groups and quantification of LAMP1/β-actin, CTSD/β-actin and expressed as fold change relative to ND group. (I) Relative lysosomal NAG activity in the livers from different groups. All experiments were repeated at least 3 times. n = 5 mice per group for A–D; n = 6 mice per group for E–H; n = 9 mice per group for I. Data were expressed as the mean ± SEM; ∗P < 0.05, ∗∗P < 0.01.