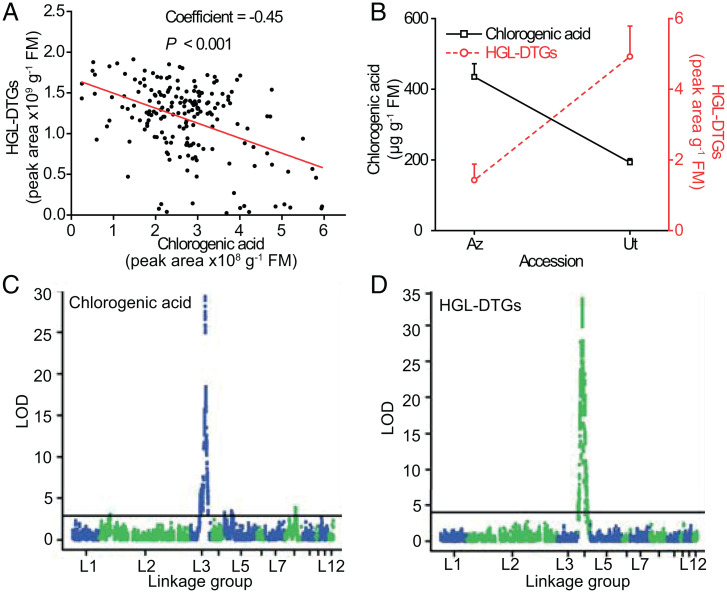
Fig. 4.
The abundances of CA and HGL-DTGs are genetically negatively correlated in natural accessions of N. attenuata and impute different loci in a RIL population. (A) Relative abundances of CA and HGL-DTGs in leaves of 183 natural accessions of N. attenuata are negatively correlated. (B) Abundances (mean + SE, n = 5) of CA and HGL-DTGs in two well-studied accessions of N. attenuata, Ut and Az. The QTL mapping using an AI-RIL population generated from intercrosses of the Ut and Az accessions and subsequent inbreeding revealed separate loci regulating the abundances of CA (C) and HGL-DTGs (D). Three genes imputed in the CA QTL—namely, a MYBb308 transcription factor (TF), a PAL biosynthetic gene, and a bHLH TF—underwent VIGS, but only the MYB308 TF significantly altered CA contents (SI Appendix, Fig. S6).