Fig. 2.
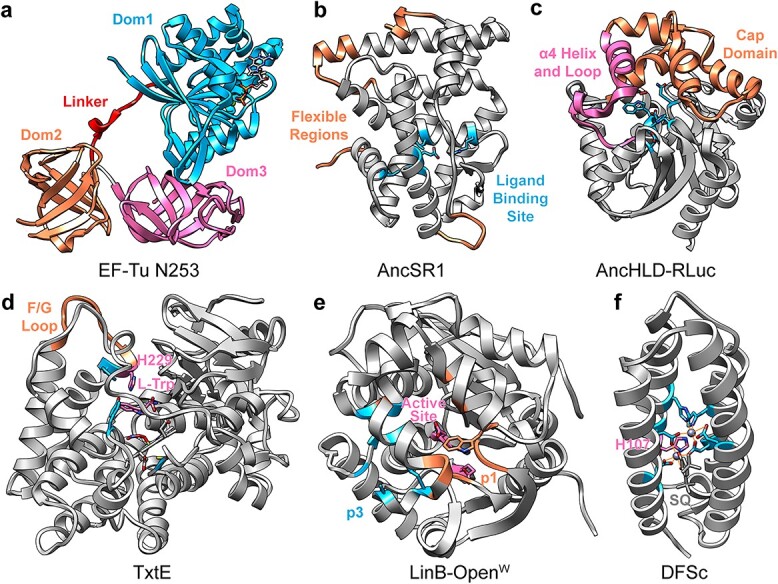
Flexible regions and stabilizing features arising from consensus designs and ancestral sequence reconstructions. (a) ASR EF-Tu N253 with GDP and Mg2+ (PDB 5w76) colored by domain with the flexible linker shown in red. (b) Model of AncSR1 (personal communication) showing the most dynamic regions (residues 1–7, 28–35, 155–168 and 245–249) and ligand-binding site (Ala47, Glu50, Leu84, Arg91 and Leu237). (c) AncHLD-RLuc (PDB 6g75) with catalytic residues (Asn51, Asp118, Trp119, Glu142 and His284), O2 substrate, α4 helix and adjacent loop (residues 142–167) and cap domain (residues168–222) shown. (d) Closed-lid TxtE (MD structure (Dodani et al., 2016)) with F/G loop (residues 175–186), wild type His229, active site residues (Tyr89, Tyr175, Cys357), L-Trp substrate and heme cofactor shown. (e) LinB-OpenW (PDB 5lka) with the native, blocked p1 tunnel (Leu177Trp), designed p3 tunnel (Trp140Ala, Phe143Leu and Ile211Leu) and active site (residues Asp108 and Asp132) shown. (f) Model of DFSc bound to semiquinone (personal communication) with Zn2+-coordinating residues (Glu11, Glu44, Glu74, His77, His100 and Glu104) and His107 shown.
