SUMMARY
TET1 maintains hypomethylation at bivalent promoters through its catalytic activity in embryonic stem cells (ESCs). However, TET1 catalytic activity-independent function in regulating bivalent genes is not well understood. Using a proteomics approach, we map the TET1 interactome in ESCs and identify PSPC1 as a TET1 partner. Genome-wide location analysis reveals that PSPC1 functionally associates with TET1 and Polycomb repressive complex-2 (PRC2). We establish that PSPC1 and TET1 repress, and the lncRNA Neat1 activates, bivalent gene expression. In ESCs, Neat1 is preferentially bound to PSPC1 alongside its PRC2 association at bivalent promoters. During the ESC-to-epiblast-like stem cell (EpiLC) transition, PSPC1 and TET1 maintain PRC2 chromatin occupancy at bivalent gene promoters, while Neat1 facilitates the activation of certain bivalent genes by promoting PRC2 binding to their mRNAs. Our study demonstrates a TET1-PSPC1-Neat1 molecular axis that modulates PRC2-binding affinity to chromatin and bivalent gene transcripts in controlling stem cell bivalency
Graphical Abstract
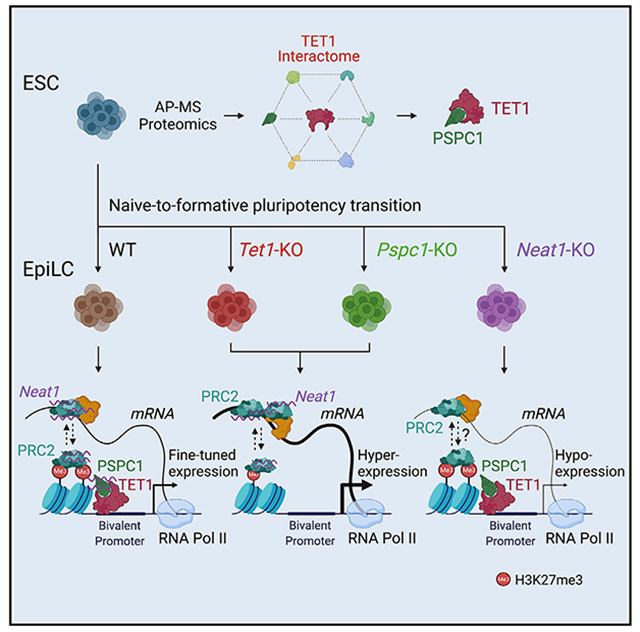
In brief
Huang et al. use proteomics and genetic approaches to show that catalytic activity-independent functions of TET1, coordinated with the paraspeckle components PSPC1 and its cognate lncRNA Neat1, dynamically regulate stem cell bivalency by modulating PRC2 binding to chromatin and bivalent gene transcripts in the naive-to-formative pluripotent state transition.
INTRODUCTION
Embryonic stem cells (ESCs) and epiblast stem cells (EpiSCs) of the naive and primed pluripotency states, respectively, differ significantly in their transcriptomic features, clonogenicity, and differentiation potentials (Nichols and Smith, 2009). Epiblast-like stem cells (EpiLCs), a kind of formative pluripotent cells, transiently emerge when adapting ESCs to primed EpiSCs culture conditions within a specific period (usually 48 h), while an extended culture of EpiLCs establishes a stable primed state (Hayashi et al., 2011; Morgani et al., 2017; Smith, 2017). Recently, stable cell lines of formative pluripotency state were generated with specific combinations of cytokines and inhibitors (Kinoshita et al., 2021; Wang et al., 2021; Yu et al., 2021) with a notable molecular feature, i.e., the “super-bivalency” at lineage-specific genes present both in vivo (Xiang et al., 2020) and in vitro (Wang et al., 2021). Bivalent promoters are marked by H3K4me3 and H3K27me3 (Bernstein et al., 2006), catalyzed by KMT2B and polycomb repressive complex-2 (PRC2), respectively, and are considered to poise the expression of developmental regulators in ESCs while allowing timely activation upon differentiation cues (Voigt et al., 2013). DNA methylation at bivalent promoters decreases KMT2B activity and H3K4me3, which in turn leads to increased PRC2 occupancy at promoters (Mas et al., 2018). The TET (ten-eleven translocation) family of proteins regulate gene expression through DNA demethylation (Kohli and Zhang, 2013), and were thus implicated in regulating bivalency (Mas et al., 2018; Xiang et al., 2020). Although the loss of TET proteins (Tet1KO or Tet1/2/3TKO) causes global changes in the DNA methylation and gene expression in ESCs, the cells nevertheless retain the ability to self-renew (Dawlaty et al., 2011; Lu et al., 2014; Verma et al., 2018). In the formative EpiLCs and the primed EpiSCs, TET1 is the only expressed TET protein (Fidalgo et al., 2016; Khoueiry et al., 2017). Loss of TET1 causes dysregulation of gene expression in ESC differentiation (Dawlaty et al., 2011; Koh et al., 2011) and defects in mouse post-implantation development (Khoueiry et al., 2017). Notably, TET1 is responsible for maintaining the DNA methylation valleys at promoters of developmentally regulated genes to establish a super-bivalency in the post-implantation epiblast (Xiang et al., 2020). Mechanistically, TET1 activates and represses gene transcription by catalytic activity-dependent and independent functions through promoter/enhancer demethylation (Kohli and Zhang, 2013) and association with SIN3A/HDAC (Williams et al., 2011) or PRC2 (Chrysanthou et al., 2022; Neri et al., 2013; Wu et al., 2011) complexes, respectively.
The post-transcriptional gene regulation by PRC2 has been increasingly appreciated through its association with RNA-binding proteins (RBPs) and long noncoding RNAs (lncRNAs) that can regulate gene expression in cis or in trans (Cifuentes-Rojas et al., 2014; Davidovich and Cech, 2015; Kaneko et al., 2014; Yan et al., 2019). In addition, nascent mRNAs and other RNA transcripts were also proposed to antagonize the association of PRC2 with the chromatin (Beltran et al., 2016; Davidovich et al., 2015; Kaneko et al., 2013; Long et al., 2020; Wang et al., 2017b). In vivo, a “PRC2 eviction” model was proposed in which the nascent mRNA regulates its own production by evicting PRC2 from the promoter, thereby further promoting gene transcription (Skalska et al., 2021; Wang et al., 2017a). Although TET1 is a putative RBP (He et al., 2016), whether/how TET1 may functionally connect with PRC2 through other RBPs and/or lncRNAs to control bivalent genes in pluripotent states has not been determined.
By studying the TET1 interactome in mouse ESCs, we here report the discovery of paraspeckle component 1 (PSPC1), a RBP generally associated with nuclear paraspeckles (Knott et al., 2016), as a TET1 partner. We further establish that PSPC1 and its cognate lncRNA Neat1 associate with TET1 and PRC2 at bivalent promoters. Using genetic loss-of-function approaches, we demonstrate that TET1 and PSPC1 promote PRC2 chromatin occupancy through Neat1 to counteract the binding of PRC2 to bivalent gene transcripts, thereby preventing PRC2 eviction from chromatin to maintain the super-bivalency during the ESC-to-EpiLC transition. On the other hand, upon the loss of TET1 or PSPC1, Neat1 enhances PRC2 binding to mRNAs, thereby activating transcription of bivalent genes during pluripotent-state transition. Our study thus establishes a previously unappreciated TET1-PSPC1-Neat1 molecular axis that modulates PRC2 occupancy at chromatin and bivalent gene transcripts in controlling stem cell bivalency.
RESULTS
The TET1 interactome in ESCs identifies PSPC1 as its interacting partner
We engineered mouse ESCs expressing FLAG-tagged TET1 (FL-Tet1) and purified the TET1 protein complexes using SILAC (stable isotope labeling by amino acid in cell culture)-based AP-MS (affinity purification followed by mass spectrometry) method as described in our previous studies (Ding et al., 2015; Guallar et al., 2018; Huang et al., 2021). Reciprocal SILAC labeling was performed as biological replicates (Rep1/2), and the intensity ratios of TET1 versus control immunoprecipitation (IP) (Rep1: light/heavy; Rep2: heavy/light) for each protein were plotted (Figure 1A; Table S1). Validating our approach, we identified several known TET1 partners such as OGT and SIN3A (Vella et al., 2013) and components of a ribosome biogenesis complex consisting of PELP1, TEX10, WDR18, and SENP3 (Finkbeiner et al., 2011), consistent with our previous finding that TET1 and TEX10 are close partners (Ding et al., 2015). In addition, we identified several RBPs such as L1TD1 and PSPC1 (Figures 1A and S1A). Selected candidate proteins in the TET1 interactome were validated by FLAG co-immunoprecipitation (co-IP) followed by western blot analysis (Figure 1B). We decided to focus on the TET1 and PSPC1 partnership for several reasons. First, although the functional significance of the TET1-SIN3A/OGT (Deplus et al., 2013; Vella et al., 2013; Williams et al., 2011) and TET1-TEX10 (Ding et al., 2015) partnerships in ESC maintenance or differentiation is well studied, the functional cooperation between TET1 and PSPC1 is unclear. PSPC1 does interact with other paraspeckle components such as SFPQ and NONO in ESCs (Figure S1B), although paraspeckles were not observed in mouse (Figure S1C) or human (Chen and Carmichael, 2009) ESCs. Second, TET1 activates and represses lineage gene expression during ESC differentiation through its catalytic activity-dependent and -independent functions, respectively (Koh et al., 2011; Wu et al., 2011; Zhu et al., 2018). Whereas TET1 catalytic activity-independent function may act through PRC2, their direct physical association was not detected (Wu et al., 2011), raising the possibility of unknown bridging proteins and/or RNAs for the functional interaction between TET1 and PRC2.
Figure 1. PSPC1 is an interacting partner of TET1 in ESCs.
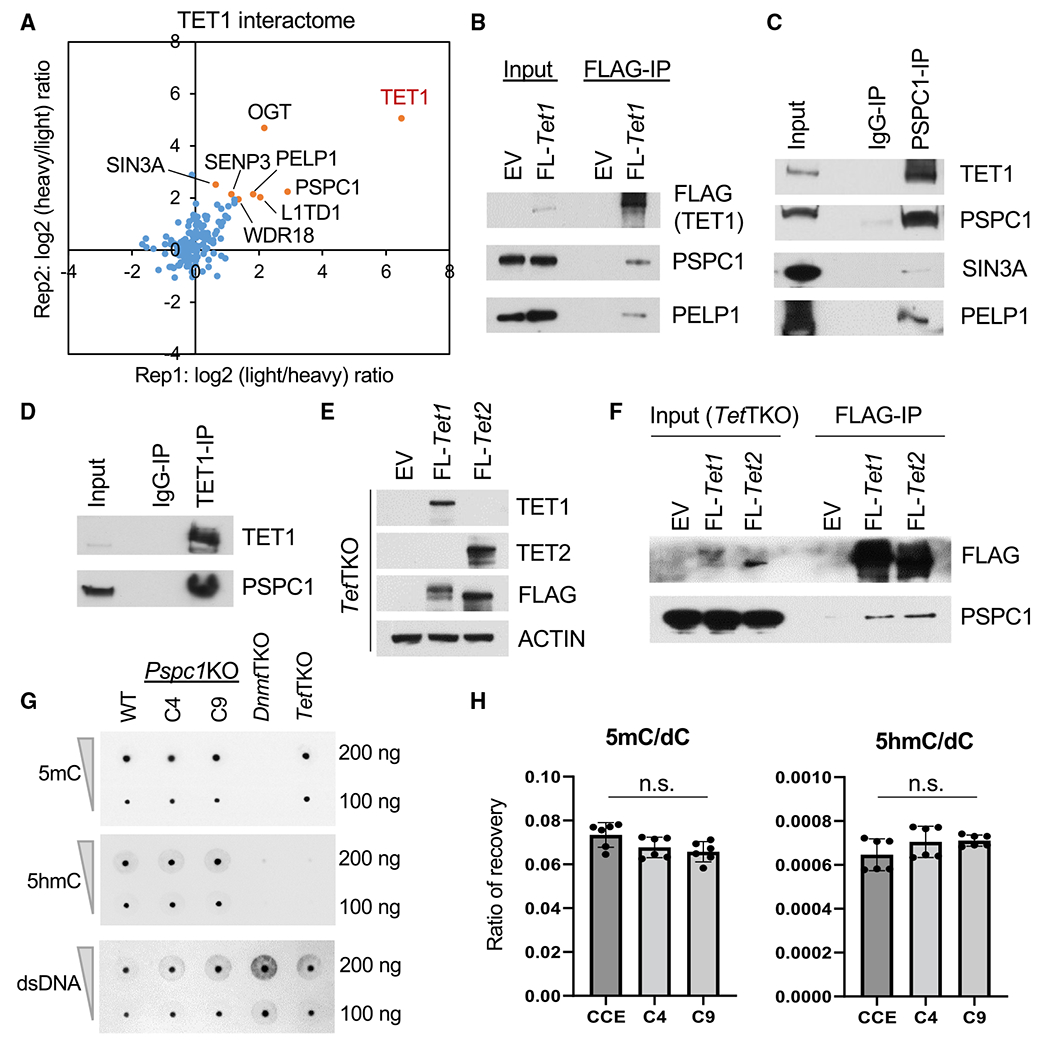
(A) Protein ratios of FLAG-IP (TET1)versus Control-IP (empty vector) AP-MS in two replicates with reciprocal SILAC labeling are plotted, and a few proteins in the TET1 interactome are indicated.
(B and F) Co-immunoprecipitation (co-IP) of TET1 partners (B) or TET1/2 (F) by FLAG-IP followed by Western blot analysis in ESCs.
(C and D) Co-IP by endogenous PSPC1 (C) and TET1 (D) antibodies followed by western blot analysis in ESCs.
(E) Western blot analysis in Tet1/2/3 triple-KO (TetTKO) ESCs rescued with FLAG-tagged TET1 or TET2 in ESCs.
(G) DNA 5mC and 5hmC dot-blot analysis of WT and Pspc1KO (two independent clones, C4 and C9) ESCs. dsDNA antibody is reblotted as the loading control. Dnmt1/3a/3b triple-KO (DnmtTKO) and TetTKO ESCs serve as negative controls of 5mC and 5hmC, respectively.
(H) UHPLC-MS/MS quantification of 5′-methyl-deoxycytidine (5mC) and 5′-hydroxymethyl-deoxycytidine (5hmC) over deoxycytidine (dC) from genomic DNA of WT and Pspc1KO ESCs. Experiments were performed in biological duplicates with technical triplicates; p value is from two-tailed t test, and “n.s.” denotes statistically non-significant.
We confirmed the interaction between PSPC1 and TET1 by reciprocal co-IP using endogenous antibodies (Figures 1C and 1D), which were independent of RNAs (Figure S1E). PSPC1 also interacts with the TET1 partners SIN3A and PELP1 (Figure 1C). Compared with the TET2 interactome, we constructed with a similar SILAC strategy in ESCs (Guallar et al., 2018), and we found that SIN3A, PSPC1, OGT, and LMNB1 are shared proteins in both interactomes. However, the proteins in the ribosome biogenesis complex (PELP1, TEX10, WDR18, and LAS1L) are present only in the TET1 interactome (Figure S1F). To probe the potential biochemical entities associated with PSPC1, TET1, and TET2, we performed size exclusion chromatography (i.e., gel filtration) on ESC nuclear extracts. We found the co-fractionation of all these three factors (complex I, blue; Figure S1G) and the TET1-free co-fractionation of PSPC1 and TET2 (complex II, red; Figure S1G). While the existent TET1-free TET2/PSPC1 complex has been demonstrated with the critical role of TET2 in RNA-dependent targeting for ERV control in ESCs (Guallar et al., 2018), we wondered whether the PSPC1-TET1 interaction is mediated by TET2 in light of their co-fractionation as seen in complex I (Figure S1G). We thus employed the Tet1/2/3 triple-KO (TetTKO) ESCs (Fidalgo et al., 2016), rescued with either FLAG-tagged TET1 or TET2 (Figure 1E; TET3 is not expressed in ESCs), and performed FLAG-IP followed by western blot of PSPC1. Interestingly, we found that both TET1 and TET2 interact with PSPC1 in the absence of the other TET proteins (Figure 1F), indicating the TET2-independent TET1-PSPC1 interaction while further confirming the TET1-independent TET2-PSPC1 interaction despite the co-fractionation of these three proteins in size exclusion chromatography.
We also performed domain-mapping experiments to dissect the TET1-PSPC1 interaction. The full-length (2,039 amino acids) or truncated fragments of Tet1 were cloned into the FLAG-tagged expression vectors (Figure S1H) for transfection in ESCs followed by Co-IP. We observed that full-length TET1 and its variants (C1, C2, and ΔCXXC) containing a minimal C-terminal catalytic domain (amino acids 1,367–2,039) interact with PSPC1 (Figure S1H). Similarly, we cloned the full-length or truncated fragments of Pspc1 into the V5-tagged expression vectors (Figure S1I) for transfection in ESCs followed by Co-IP. We found that full-length PSPC1 and its truncated variant F2 containing the multifunctional Drosophila behavior/human splicing (DBHS) domain (Knott et al., 2016) were required to interact with TET1 (Figure S1I). We then asked whether PSPC1 can modulate catalytic activity-dependent or independent functions of TET1 in ESCs. We employed Pspc1KO ESCs (two independent clones, C4 and C9, shown in Figure S1D) (Guallar et al., 2018) and performed DNA dot-blot and mass spectrometry analysis. We found that PSPC1 ablation does not affect the DNA 5mC or 5hmC intensity in ESCs (Figures 1G and 1H). Taking these together, we identified PSPC1 as a TET1 partner that may modulate TET1 functions in ESC pluripotency independently of its catalytic activity.
PSPC1, TET1, and PRC2 co-localize at the bivalent gene promoters in ESCs
PSPC1 is a DNA- and RNA-binding protein (Knott et al., 2016). Therefore, we performed chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) analysis of PSPC1 in WT and Pspc1KO ESCs. We identified 2,324 PSPC1 ChIP-seq peaks in ESCs, using PSPC1 ChIP in Pspc1KO cells as the background control. The majority (74.2%) of PSPC1-binding peaks are located at the gene promoters (within 5K bp of transcriptional start sites, TSSs), with PSPC1 ChIP signal also enriched at TSSs (Figures 2A and 2B). Consistent with the PSPC1-TET1 partnership, almost all PSPC1 peaks (91.7%, 2,132/2,324) co-localize with TET1-binding regions (Figure S2A). We compared the DNA 5hmC and 5mC intensities at the TET1 peak regions with or without PSPC1 occupancy from published (hydroxy)methylated DNA immunoprecipitation sequencing (hme/meDIP-seq) data in ESCs (Xiong et al., 2016). Overall, the PSPC1/TET1 common regions lack 5hmC and 5mC compared with the TET1-only regions (Figure S2B), consistent with our finding that PSPC1 does not participate in the catalytic activity-dependent functions of TET1 in ESCs (Figures 1G and 1H).
Figure 2. PSPC1, TET1, and PRC2 co-localize at bivalent promoters in ESCs.
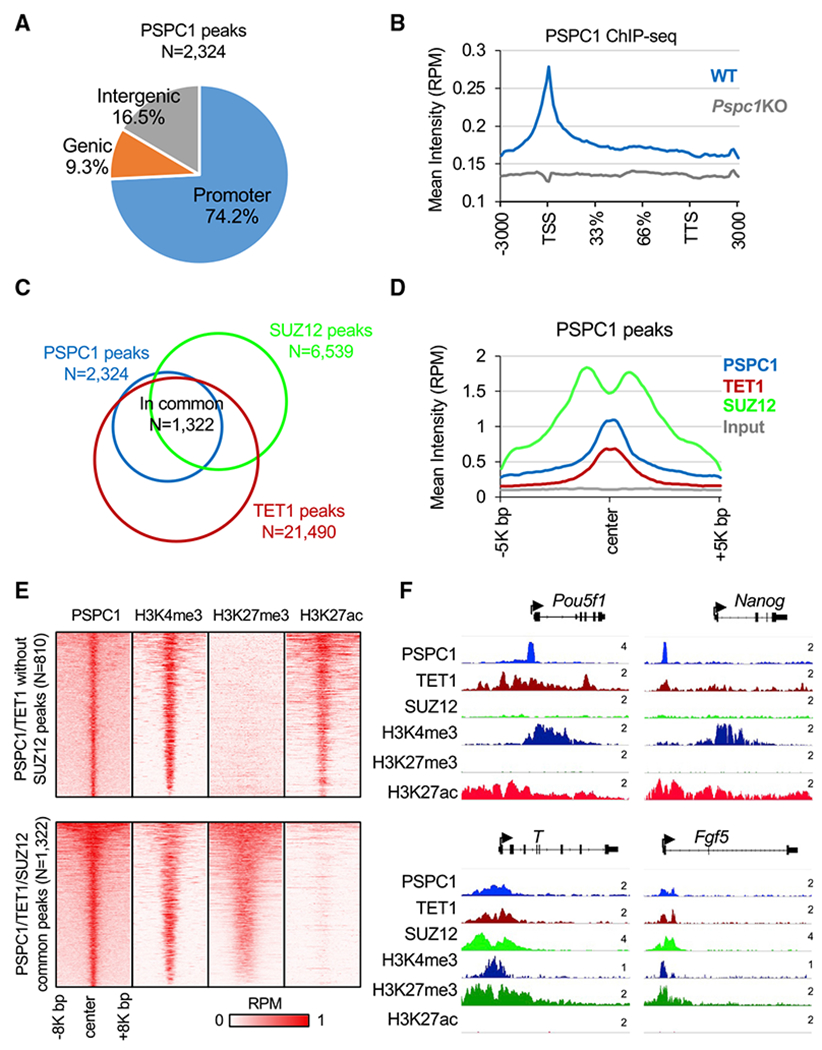
(A) Annotation of PSPC1 ChIP-seq peaks in ESCs at promoters, intergenic or genic regions.
(B) Mean intensity plot by reads per million (RPM) showing PSPC1 ChIP-seq intensity of WT and Pspc1KO ESCs at gene bodies (within 3K bp). TSS, transcription start site, TTS, transcription termination site.
(C) Overlap ofthe PSPC1, PRC2 subunit SUZ12, and TET1 (Wu et al., 2011) peaks in ESCs.
(D) Mean intensity plot by RPM showing PSPC1, TET1, and RPC2 subunit SUZ12 ChIP-seq intensity at PSPC1 peak regions (within 5K bp at PSPC1 peak center).
(E) Heatmaps by RPM showing PSPC1 and histone marks H3K4me3, H3K27ac (Hon et al., 2014), and H3K27me3 (Cruz-Molina et al., 2017) at PSPC1/TET1 common peak regions (within 8K bp at PSPC1 peak center) with and without PRC2 occupancy.
(F) ChIP-seq tracks of PSPC1, TET1, SUZ12, and histone marks of H3K4me3, H3K27me3, and H3K27ac at PSPC1/TET1 common peak regions with (T, Fgf5) and without (Pou5f1, Nanog) PRC2 occupancy. The numbers indicate the normalized RPM value of the tracks.
To understand how PSPC1 may functionally interact with other transcriptional regulators in ESCs, we performed ChIP-seq correlation analysis (Ding et al., 2015) and found that PSPC1 DNA binding sites are more like those of TET1 and EZH2/SUZ12 (Figure S2C), suggesting that PSPC1 may be involved in TET1- and PRC2-dependent regulations. Indeed, 56.9% (1,322/2,324) of the PSPC1 peaks are co-occupied by TET1 and PRC2 component SUZ12 (Figure 2C). TET1 and SUZ12 are also enriched at PSPC1-bound regions (Figure 2D). PRC2 deposits the repressive histone mark H3K27me3 at the promoters of bivalent genes in ESCs that are lowly expressed and poised to be promptly activated upon differentiation (Boyer et al., 2006). Consistently, gene ontology (GO) analysis for the PSPC1/TET1/SUZ12 common targets revealed that many of the genes are involved in multicellular organism development, cell fate commitment, and cell differentiation (Figure S2D). Next, we compared the intensity of histone marks H3K4me3, H3K27ac,and H3K27me3 at the PSPC1/TET1 common peaks with or without SUZ12 occupancy. The PSPC1/TET1 peaks without SUZ12 occupancy were enriched with active marks of H3K4me3 and H3K27ac (e.g., promoters of Pou5f1 and Nanog), whereas the PSPC1/TET1/SUZ12 common peaks were enriched with bivalent marks of H3K4me3 and H3K27me3 (e.g., promoters of T and Fgf5) (Figures 2E and 2F). RNA-seq analysis of Pspc1KO ESCs (this study) or Tet1KO ESCs (Hon et al., 2014) indicated that depletion of PSPC1 or TET1 protein does not disturb the expression of PSPC1/TET1 common target genes (with or without SUZ12 occupancy) (Figure S2E), consistent with the fact that Pspc1KO (Guallar et al., 2018) or Tet1KO (Hon et al., 2014) does not affect the maintenance of ESCs.
Together, these results suggest a potential physical association of the TET1-PSPC1 partnership with PRC2 in repressing bivalent genes in ESCs. However, the possible role of the TET1-PSPC1 partnership independent of PRC2 in activating pluripotency genes cannot be discounted and warrants future investigation (see Discussion).
PSPC1 restricts bivalent gene activation during the ESC-to-EpiLC transition
To understand how PSPC1 might contribute to the regulation of bivalent genes in pluripotent cells, we decided to study the functions of PSPC1 in the pluripotent-state transition, during which the super-bivalency of a large set of developmental genes was initially proposed (Morgani et al., 2017; Smith, 2017) and subsequently confirmed (Wang et al., 2021) in formative pluripotent stem cells. By switching the culture medium from serum/LIF to Fgf2/activin A (FA), ESCs enter a transient formative pluripotency state of EpiLCs, followed by a primed pluripotency state of EpiSCs under an extended culture of EpiLCs in the FA condition (Smith, 2017). We thus adapted WT and Pspc1KO ESCs (day 0, D0) in FA culture medium for 2 days (D2) and 4 days (D4) and collected RNAs for RNA-seq analysis (Figure 3A). Of note, the D2 EpiLCs are considered as the state of formative pluripotency (Buecker et al., 2014; Fidalgo et al., 2016; Hayashi et al., 2011), whereas D4 EpiLCs and EpiSCs are of primed pluripotency when the meso/ectodermal lineage genes (e.g., Fgf5, Fgf8, T, Eomes, and Otx2) are further activated (Huang et al., 2017). Principal-component analysis (PCA) revealed a trajectory of gene expression profiles moving from D0 (ESC) to D2 and D4 (EpiLC) on PC1, while the differences of gene expression between WT and KO cells at all three time points are reflected on PC2 (Figure 3B). By comparing the differentially expressed genes (DEGs; p < 0.05, fold change > 1.5; Table S2) between WT and Pspc1KO cells at three time points, we found that multiple signaling pathways and their associated genes, including FGF signaling (e.g., Fgf5 and Fgf8), Nodal signaling (e.g., Nodal and Eomes), and Wnt signaling (e.g., Axin2, Wnt5b, and Wnt8a), are upregulated in Pspc1KO relative to WT EpiLCs (D2 and D4; Figure S3A). GO analysis of these PSPC1-repressed DEGs in D2 and D4 EpiLCs indicates that they are involved in embryo and tissue development (Figure S3B, left). In contrast, the PSPC1-activated DEGs are involved in multiple cellular regulations, including metabolic process, protein transport, and cell death (Figure S3B, right). Interestingly, a majority (75.9%, 129/170) of the PSPC1-repressed DEGs in EpiLCs are not repressed by PSPC1 in ESCs (Figure S3B, left), likely due to their low expression levels and/or alternative repression mechanisms in ESCs.
Figure 3. PSPC1 negatively regulates activation of bivalent genes in the pluripotent-state transition.
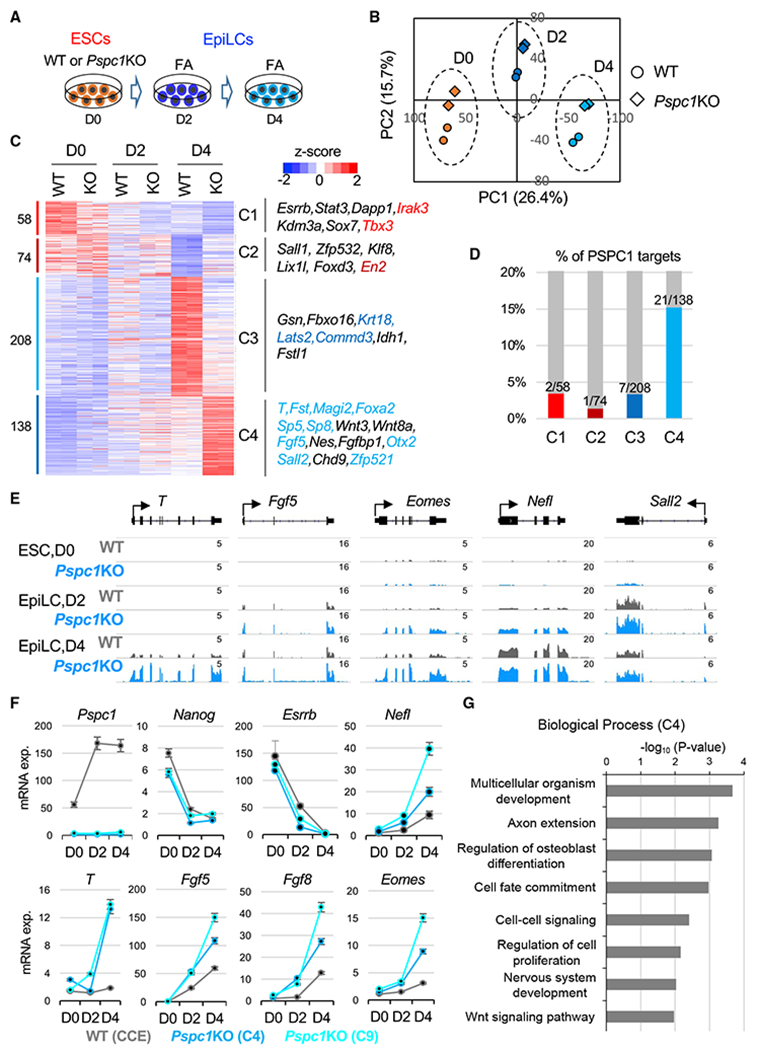
(A) Schematic depiction of the naive-to-formative transition of WT and Pspc1KO ESCs. The ESCs are adapted in Fgf2 and activin A (FA) culture medium for 2 days and 4 days.
(B) Principal-component analysis (PCA) of WT and Pspc1KO RNA-seq samples at different time points. Percentages of variance explained in each principal component (PC) are indicated.
(C) Heatmap showing the relative expression of differentially expressed genes (DEGs) by comparing D0 WT with D4 WT cells and D4 WT with D4 KO cells. The numbers of DEGs are shown on the left, and representative genes in the four classes (C1–C4) are listed on the right. The direct PSPC1 targets from ChIP-seq analysis are indicated by the color text, which matches the color of the histogram in (D).
(D) Histogram showing the percentages (%) and numbers of DEGs in each class (C1–C4) as the PSPC1 ChIP-seq targets.
(E) RNA-seq tracks of WT and Pspc1KO ESCs and EpiLCs at bivalent lineage gene loci. The numbers indicate the normalized RPM value of the tracks.
(F) RT-qPCR analysis of pluripotency and lineage genes in WT and Pspc1KO ESCs (CCE background with two independent clones, C4 and C9) during ESC-to-EpiLC differentiation. Error bars represent the standard deviation of technical triplicates.
(G) Gene ontology (GO) analysis for the C4 genes (Class 4, N = 138) shown in (C).
Next, we focused on the DEGs between D0 and D4 (ESC vs. EpiLC) WT cells and between D4 WT and Pspc1KO EpiLCs to obtain 478 shared DEGs (Figure 3C; Table S2). Clustering analysis of these genes illustrated different expression patterns among the samples (class 1–4, or C1–4; Figure 3C). We examined the number of DEGs in C1–4 that were direct PSPC1 targets from ChIP-seq analysis and found that C4 contains the highest percentage (15.2%, 21/138) of PSPC1 targets (Figure 3D). These PSPC1 targets (e.g., T, Fgf5, and Sall2) are bivalent and minimally expressed in ESCs, while transcriptionally activated in EpiLCs, and PSPC1 depletion further increases their expression during EpiLC differentiation (Figures 3C, 3E, and 3F). GO analysis of these PSPC1-repressed C4 genes indicated that they were involved in multicellular organism development, cell fate commitment, and Wnt-signaling pathways (Figure 3G). To examine whether the repressive effect of PSPC1 on bivalent gene expression is dependent on its RNA-binding capacity, we rescued Pspc1KO ESCs with either a PSPC1 WT or an RNA recognition motif mutant (RRMmut) protein and performed EpiLC differentiation. Our data revealed that only the WT, but not the RRMmut protein, could rescue the repressive effect of PSPC1 on the target genes (e.g., Fgf5, Fgf8, and Wnt8a) (Figures S3C and S3D). Of note, NONO, a close partner of PSPC1 (Figure S1B), also interacts with TET1 in ESCs (Li et al., 2020). Like Pspc1KO, NonoKO is compatible with ESC maintenance (Ma et al., 2016) and causes upregulation of lineage genes (e.g., Eomes, Fgf5, Fgf8, and T) in EpiLCs (Figures S3E and S3F). In sum, our results establish PSPC1 as a transcriptional repressor that restricts bivalent gene activation during the ESC-to-EpiLC transition.
Neat1 promotes bivalent gene activation during the ESC-to-EpiLC transition
PSPC1 as an RBP was well known for its roles in binding lncRNA Neat1, which drives the formation of nuclear paraspeckles (Isobe et al., 2020; Nakagawa et al., 2011). However, pluripotent stem cells do not form paraspeckles, and thus the functional relationship between PSPC1 and Neat1 in pluripotency is not fully understood. Neither is it known whether Neat1 plays any role in modulating TET1 functions. Therefore, we designed two sgRNAs targeting the Neat1 locus and performed CRISPR-Cas9 genome editing to delete the 6K-bp region containing the short (Neat1_1) isoform of Neat1 (Figure 4A), the only isoform expressed in ESCs (Isobe et al., 2020) and EpiLCs (Figure 4B). Of note, the long Neat1_2 is a somatic isoform that functions in driving paraspeckle formation (Isobe et al., 2020) and is collaterally abrogated by our CRISPR deletion (Figure 4A). We thus collectively refer to Neat1KO hereafter. We adapted WT and Neat1KO ESCs in FA culture medium and collected RNAs at D0 (ESC), D2, and D4 (EpiLC) for RNA-seq analysis. Like Pspc1KO ESCs, Neat1KO ESCs (two independent clones, 5F and 7G) are properly maintained with unaltered protein and mRNA expression of representative pluripotency/lineage genes critical for ESC maintenance/differentiation (Figures S4A and S4B). We confirmed that only Neat1_1 (the short isoform) is expressed in ESCs and EpiLCs, and its expression gradually decreases during EpiLC differentiation (Figure 4B; reduced signal strengths on the left panel and FPKM values on the right panel). By comparing the DEGs (p < 0.05, fold-change > 1.5; Table S3) between the WT and Neat1KO EpiLCs, we observed many bivalent genes (e.g., Fgf5, Fgf8, Nefl, and Wnt8a) are downregulated in D2 or D4 EpiLCs upon Neat1KO compared with the WT cells (Figure S4C), confirmed by quantitative PCR (qPCR) analysis (Figure 4E). Interestingly, the effect of Neat1KO on bivalent genes (Figures 4D, 4E, and S4C) is opposite to that of Pspc1KO (Figures 3E, 3F, and S3A) in EpiLCs. Fewer DEGs were identified in D4 EpiLCs relative to D0 ESCs and D2 EpiLCs upon Neat1KO (Figure S4C), likely due to the relatively low Neat1 expression in D4 EpiLCs (Figure 4B).
Figure 4. Neat1 positively regulates bivalent gene activation in the pluripotent-state transition.
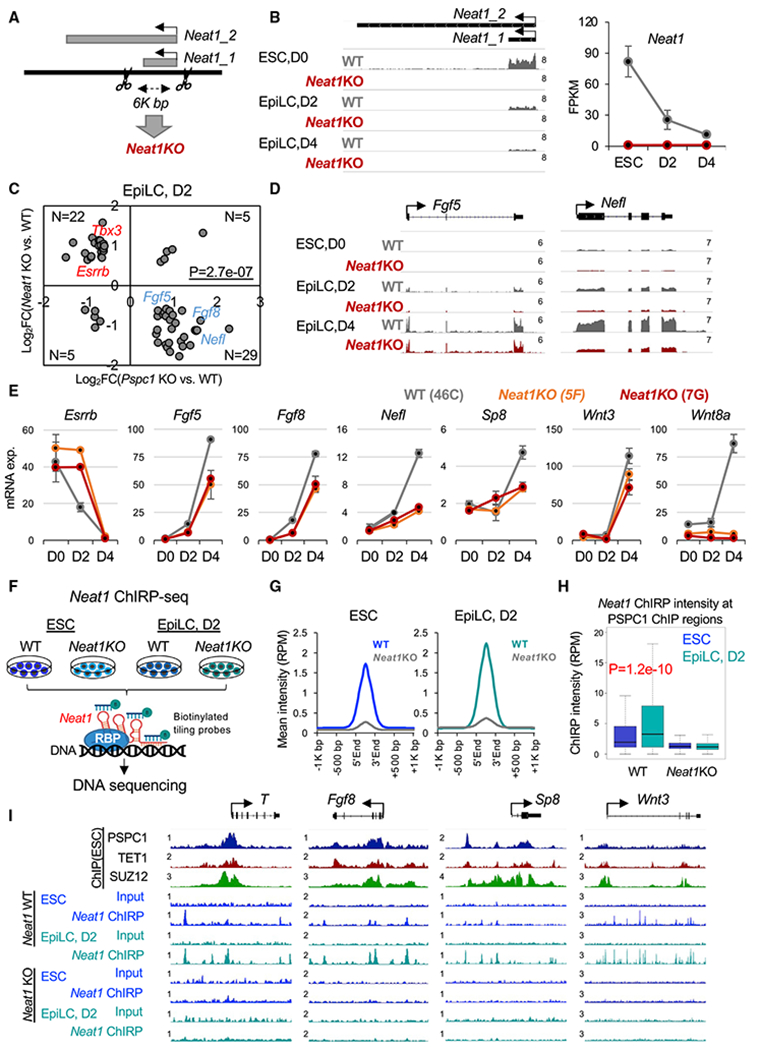
(A) Schematic depiction of the Neat1KO strategy. The scissors denote two gRNA-targeting sites for CRISPR-Cas9 genome editing. The short (Neat1_1) and long (Neat1_2) isoforms of the mouse Neat1 gene are indicated.
(B) RNA-seq tracks (left) and expression of Neat1 (right) during the ESC-to-EpiLC differentiation. The numbers indicate the normalized RPM value of the tracks (left). Neat1 expression is shown in FPKM (fragments per kilobase of transcript per million mapped reads) values (right). Error bars represent the standard deviation of biological duplicates.
(C) Scatter plot of the relative gene expression of DEGs upon Pspc1KO or Neat1KO relative to WT at D2 EpiLC from RNA-seq analysis; p value is from the Fisher exact test. Representative genes are labeled on the plot.
(D) RNA-seq tracks of WT and Neat1KO ESCs and EpiLCs at bivalent gene loci (Fgf5 and Nefl). The numbers indicate the normalized RPM value of the tracks.
(E) RT-qPCR analysis of bivalent genes in WT and Neat1KO ESCs (46C genetic background with two independent clones, 5F and 7G) during EpiLC differentiation. Error bars represent the standard deviation of technical triplicates.
(F) Schematic depiction of Neat1 ChIRP-seq analysis in WT and Neat1KO ESCs and D2 EpiLCs. Biotinylated probes based on their relative positions along the Neat1_1 RNA were ranked and split into odd and even probes, followed by streptavidin pull-down and DNA sequencing.
(G) Mean intensity plot by RPM showing Neat1 ChIRP-seq intensity enriched at the Neat1 peak regions in ESCs and D2 EpiLCs (within 1K bp around Neat1 peak regions identified in ESCs).
(H) Boxplots depicting quantification of Neat1 ChIRP-seq intensity by RPM at PSPC1 ChIP-seq peak regions (extend 5K bp, identified in ESCs) from WT and Neat1KO ESCs and D2 EpiLCs. p value is from the Mann-Whitney test.
(I) PSPC1, TET1, and SUZ12 ChIP-seq tracks in ESCs and Neat1 ChIRP-seq tracks in WT and Neat1KO ESCs and D2 EpiLCs at the promoters of bivalent genes (T, Fgf8, Sp8, and Wnt3). The numbers indicate the normalized RPM value of the tracks.
To further investigate the functional relationship between PSPC1 and Neat1, we compared the RNA-seq gene expression ratios upon Pspc1KO and Neat1KO at three time points. We again observed a negative correlation of gene expression in ESCs (r = −0.27) and D2 EpiLCs (r = −0.23), but a weak positive correlation (r = 0.08) in D4 EpiLCs (Figure S4D). Next, we plotted the gene expression ratios of DEGs by Pspc1KO and Neat1KO at different time points (Figures 4C and S4E). Interestingly, whereas Pspc1KO decreases and increases the expression of pluripotency (e.g., Esrrb and Tbx3) and bivalent (e.g., Fgf5, Fgf8, and Nefl) genes, respectively, in D2 EpiLCs, as previously observed (Figures 3 and S3), Neat1KO exhibits an opposite effect in the regulation of those genes (Figures 4C–4E and S4E). The PCA analysis of the Pspc1KO and Neat1KO RNA-seq samples shows that the D0 (ESC) and D2 and D4 (EpiLC) samples group together, indicated by dash line circles, and move rightward on PC1 during EpiLC differentiation (Figure S4F). Consistent with the correlation analysis (Figures 4C, S4D, and S4E), the Pspc1KO and Neat1KO samples deviated to opposite directions compared with their WT samples in D0 and D2 (Figure S4F; refer to the direction of red versus blue arrows at each time point).
To understand the genomic occupancy of Neat1 in ESCs and during EpiLC differentiation, we performed ChIRP (chromatin isolation by RNA purification) with split pools of tiling probes covering the Neat1_1 isoform followed by deep sequencing (ChIRP-seq) (Figure 4F). The Neat1KO cells were employed as the negative control. As expected, the Neat1 ChIRP signal significantly enriched at the Neat1 locus in both ESCs and EpiLCs (Figure S4G). In addition, we confirmed that Neat1 was highly enriched at the Sfi1 locus at chromosome 11.qA1 region (Figure S4H), a known Neat1 target site previously reported from a global survey of genome-wide lncRNA-chromatin interactions in ESCs (Bonetti et al., 2020). Interestingly, the Sfi1 locus was also co-occupied with PSPC1, TET1, and SUZ12 from the ChIP-seq data in ESCs (Figure S4H), suggesting a genomic association of Neat1 and these proteins on chromatin. When comparing the Neat1 ChIRP reads enriched at the Neat1 peak regions in ESCs and D2 EpiLCs, we observed an overall higher intensity in D2 EpiLCs than that in ESCs (Figure 4G) despite its relatively lower expression level in D2 EpiLCs than in ESCs (Figure 4B). Importantly, Neat1 ChIRP intensity at the overall PSPC1 ChIP peaks (identified in ESCs) was also significantly higher in D2 EpiLCs than ESCs (Figure 4H). For example, we observed higher Neat1 ChIRP signals at the PSPC1/SUZ12/TET1 co-occupied bivalent gene promoters (e.g., T, Fgf8, Sp8, and Wnt3) in D2 EpiLCs than in ESCs (Figure 4I). Together, our results demonstrate that Neat1 may promote bivalent gene activation through its enhanced association with bivalent chromatin during the ESC-to-EpiLC transition, establishing opposing functions of PSPC1 and its cognate lncRNA Neat1 in controlling bivalent gene expression in pluripotent-state transition.
PSPC1 is required for maintaining PRC2 chromatin occupancy and H3K27me3 deposition at bivalent promoters during the ESC-to-EpiLC transition
The opposing functions of PSPC1 and its cognate lncRNA Neat1 in controlling bivalent gene expression prompted us to examine their potential roles in modulating TET1 and PRC2 functions on transcriptional regulation of bivalent genes. We first asked whether PSPC1 contributes to TET1 and PRC2 chromatin binding. In ESCs, Pspc1KO does not affect the chromatin-bound fraction of TET1 or the PRC2 subunit SUZ12 (Figure S5A). We then addressed the potential roles of TET1 in the ESC-to-EpiLC transition. We established a degron system (Nabet et al., 2018) for rapid and inducible TET1 protein degradation (Figures 5A and 5B; two independent clones, C#13 and C#16; see details in STAR Methods). Using Tet1-degron ESCs, we confirmed that activation of lineage genes (e.g., T, Fgf5, and Fgf8) during the ESC-to-EpiLC transition is further enhanced by dTAG13 treatment (i.e., TET1 depletion) (Figure S5B), phenocopying Pspc1KO (Figure 3F).
Figure 5. Depletion of PSPC1 or TET1 accelerates PRC2 eviction from bivalent promoters.
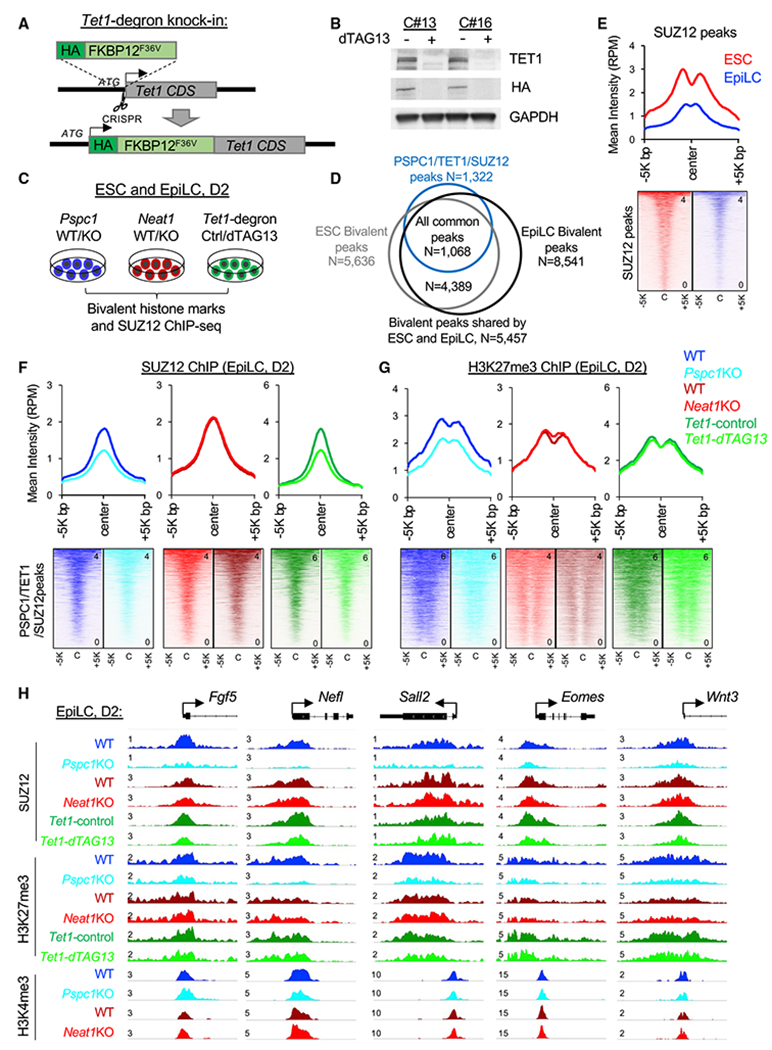
(A) Schematic depiction of the Tet1-degron knock-in (KI) strategy using CRISPR-Cas9 genome-editing tool (the scissor symbol). The HA-tagged FKBP12F36V donor sequence is inserted right after the start codon (ATG) of TET1 CDS to create the in-frame fusion protein.
(B) Western blot analysis of TET1 protein in Tet1-degron ESCs (two independent clones, C#13 and C#16) upon dTAG13 treatment for 24 h. Degradation of TET1 was indicated by both endogenous antibody and HA fusion protein tag.
(C) Schematic depiction of the bivalent histone marks H3K4me3 and H3K27me3 and the PRC2 subunit SUZ12 ChIP-seq analysis in ESCs and D2 EpiLCs of different genotypes (Pspc1 WT/KO and Neat1 WT/KO) or treatment (Tet1-degron with control/dTAG13).
(D) Overlap of the bivalent peaks (H3K4me3 and H3K27me3) identified in ESCs and EpiLCs and with the PSPC1/TET1/SUZ12 common peaks identified in ESCs.
(E) Mean intensity plot (top) and heatmap (bottom) by RPM of SUZ12 ChIP-seq intensity in WT ESCs and EpiLCs at SUZ12 peak regions (within 5K bp at peak center, identified in ESCs).
(F and G) Mean intensity plot (top) and heatmap (bottom) by RPM of SUZ12 (F) and H3K27me3 (G) ChIP-seq intensity in D2 EpiLCs at PSPC1/TET1/SUZ12 common peak regions (within 5K bp at peak center, identified in ESCs).
(H) SUZ12, H3K4me3, and H3K27me3 ChIP-seq tracks at the promoters of bivalent genes (Fgf5, Nefl, Sall2, Eomes, and Wnt3) in D2 EpiLCs. The numbers indicate the normalized RPM value of the tracks.
Next, we asked how the PSPC1-TET1 partnership and the PSPC1/Neat1 opposing functions might impose upon PRC2 and bivalent histone marks in regulating bivalent genes during the pluripotent-state transition. We performed SUZ12, H3K4me3, and H3K27me3 ChIP-seq analysis in ESCs and D2 EpiLCs of Pspc1WT/KO or Neat1WT/KO genotypes and control- or dTAG13-treated Tet1-degron cells (Figure 5C). We chose D2 EpiLCs because a high anti-correlation was observed between the Pspc1KO and Neat1KO RNA-seq data (Figure 4C), and D2 EpiLCs represent the formative state of pluripotency where super-bivalency was established (Wang et al., 2021; Xiang et al., 2020). We identified 5,636 and 8,541 bivalent peaks from ESCs and EpiLCs, respectively, and the majority peaks (N = 5,457, 96.8% of ESC peaks and 63.9% of EpiLC peaks) were shared between the two pluripotent states. Among those bivalent peaks, 1,068 (out of 1,322, 80.8%) were shared with the PSPC1/TET1/SUZ12 common peaks identified in ESCs (Figure 5D), suggesting that most of the PSPC1/TET1/SUZ12 target regions preserved bivalency during the ESC-to-EpiLC transition. As expected, PRC2 chromatin-binding intensity at SUZ12 peak regions (identified in ESCs) decreased in D2 EpiLCs compared with ESCs (Figure 5E). Plotting the SUZ12-binding intensity at SUZ12 peaks from Pspc1KO, Neat1KO, and dTAG13-treated Tet1-degron D2 EpiLCs, we found that SUZ12 binding (measured by the mean intensity in RPM) decreased upon the depletion of PSPC1 or TET1, but not Neat1, at the PSPC1/SUZ12/TET1 common peak regions (Figure 5F), exemplified by a few bivalent promoters (e.g., Fgf5, Nelf, Sall2, Eomes, and Wnt3) (Figure 5H).
Next, we investigated the effects of PSPC1, Neat1, or TET1 depletion on bivalent histone marks during the ESC-to-EpiLC transition. Whereas Pspc1KO does not change H3K4me3 deposition in ESCs or EpiLCs (Figure S5C), Pspc1KO decreases H3K27me3 deposition in D2 EpiLCs, consistent with reduced SUZ12 binding at the PSPC1/TET1/SUZ12 common regions (Figures 5F–5H). However, in ESCs, Pspc1KO slightly increased the SUZ12 chromatin binding and H3K27me3 (Figures S5D–S5F), which was opposite to the effects of PSPC1 loss on SUZ12 and H3K27me3 in D2 EpiLCs (Figures 5F and 5G). This discrepancy may be due to the expression of the bivalent genes being mostly repressed in ESCs but activated in EpiLCs (see Discussion). The Neat1KO and dTAG13-treated Tet1-degron D2 EpiLCs showed only subtle changes of H3K27me3 relative to WT and DMSO-treated control cells, respectively, at the PSPC1/TET1/SUZ12 common regions (Figure 5G). Of note, in both ESCs and D2 EpiLCs, we observed more pronounced changes of H3K27me3 upon Pspc1KO than by Neat1KO or TET1 depletion (compare the Δ[mean intensity] between Pspc1KO, Neat1KO, or Tet1-dTAG13 relative to their WT/DMSO-treated control in Figures 5G and S5D). These results suggest a closer functional partnership of PSPC1 with PRC2 in chromatin binding and H3K27me3 deposition than with Neat1 and TET1.
To understand if Neat1KO could affect PSPC1 and TET1 chromatin binding, we also performed ChIP-qPCR analysis on a few bivalent loci (e.g., Eomes, T, and Fgf5). We found that PSPC1 and TET1 ChIP signals in both ESCs and D2 EpiLCs decreased in Neat1KO relative to WT (Figure S5G). These results together demonstrate the requirement of Neat1 for bivalent chromatin occupancy of TET1 and PSPC1, which in turn maintain PRC2 chromatin occupancy and H3K27me3 deposition at bivalent promoters during the ESC-to-EpiLC transition.
PSPC1 and TET1 act through Neat1 to modulate PRC2 binding to bivalent gene transcripts and control stem cell bivalency
While a physical association between the PSPC1-TET1 partnership and PRC2 is highly speculated (Figures 2C and 2D) for the observed functional interactions among these factors, neither a previously published work (Wu et al., 2011) nor our current APMS study (Figure 1) can detect the TET1 and PRC2 interaction or the interactions between PSPC1 and PRC2 subunits using a regular nucleosome-free co-IP protocol (Figure S6A; see STAR Methods for details). However, using a nucleosome-containing co-IP protocol with micrococcal nuclease digestion of chromatin, we and others readily detected the physical associations between PSPC1 and PRC2 subunit SUZ12 (Figure 6A) and between TET1 and PRC2 (Neri et al., 2013) in ESCs, respectively, raising the possibility of nucleosomal DNA/RNA molecules for bridging the protein interactions. By examining the datasets of PRC2 subunit EZH2 PAR-CLIP-seq (photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation followed by sequencing) (Kaneko et al., 2013) and our PSPC1 CLIP-seq (Guallar et al., 2018) in ESCs, we observed that both EZH2 and PSPC1 were enriched at the Neat1 transcripts (Figure S6B). In addition, we performed a biotinylated RNA pull-down assay (Rinn et al., 2007) to identify Neat1-associated proteins in ESCs. Interestingly, we found that EZH2 bound to Neat1 sense (Neat1-S) RNA with a relatively higher affinity than the antisense (Neat1-AS) RNA. Such a preferential Neat1 sense RNA binding was even more pronounced for PSPC1 (Figure 6B). CLIP-qPCR analysis of PSPC1 and EZH2 confirmed the binding of both proteins to Neat1 transcripts in WT ESCs (Figure 6C). Importantly, we also observed an enrichment of the Neat1 ChIRP intensity at the bivalent regions in both ESCs and EpiLCs (Figure 6D). Furthermore, consistent with the nature of promiscuous RNA binding by PRC2 (Davidovich et al., 2013; Long et al., 2020), we found that EZH2 binding to Neat1 was not affected by the loss of Pspc1 in ESCs (Figure 6C) or the loss of Pspc1 or Tet1 in D2 EpiLCs (Figure S6C), suggesting that PRC2 binding to Neat1 is independent of other RBPs such as PSPC1 irrespective of pluripotent states.
Figure 6. PSPC1, TET1, and Neat1 modulate PRC2 binding to nascent bivalent gene transcripts during bivalent gene activation.
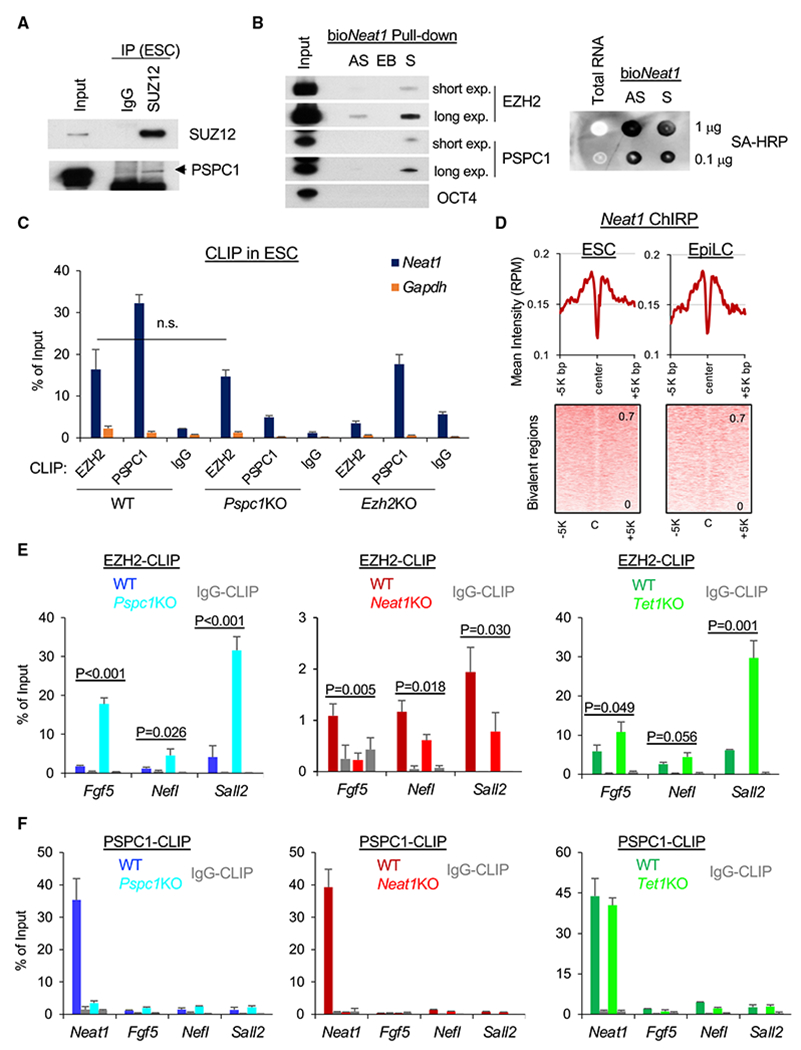
(A) Co-IP of PSPC1 and SUZ12 in ESCs using a nucleosome-containing protocol (see STAR Methods for detail).
(B) Biotinylated Neat1 (bioNeat1) RNAs pull down both EZH2 and PSPC1. Left: streptavidin (SA) beads conjugated with Neat1 sense (S) or antisense (AS) RNA, and empty beads(EB) were used for pull-down from ESC nuclear lysates followed by western blot analysis of bioNeat1-bound proteins. EZH2 and PSPC1 blots of both short and long exposure (exp.) are shown. Right: bioNeat1 sense (S) or antisense (AS) RNA were transcribed by in vitro transcription (IVT) and confirmed by SA-HRP dot blot. ESC total RNA serves as a negative control.
(C) EZH2 and PSPC1 CLIP-qPCR analysis of Neat1 in WT, Pspc1KO, and Ezh2KO ESCs. Gapdh serves as a negative control; p value is from two-tailed t test, and “n.s.” denotes statistically non-significant.
(D) Mean intensity plot (top) and heatmap (bottom) by RPM of Neat1 ChIP-seq intensity at the bivalent regions (within 5K bp at peak center, identified in ESCs) in ESCs and D2 EpiLCs.
(E) EZH2 CLIP-qPCR analysis of bivalent gene mRNAs (Fgf5, Nefl, and Sall2) in D2 EpiLCs of different genotypes (WT versus KO); p value is from the two-tailed t test.
(F) PSPC1 CLIP-qPCR analysis of Neat1 and bivalent genes’ transcripts (Fgf5, Neff, and Sall2) in D2 EpiLCs of different genotypes. Error bars in (C), (E), and (F) represent the standard deviation of technical triplicates. Experiments were repeated in biological duplicates.
Since PRC2 has a higher affinity to RNA than to DNA or histone, the nascent mRNAs during transcription activation decoy PRC2 and promote PRC2 eviction from chromatin (Wang et al., 2017a, 2017b). We hypothesized that the TET1-PSPC1-Neat1 molecular interplay might modulate PRC2 binding to nascent bivalent gene transcripts in controlling stem cell bivalency. To address this, we performed EZH2 CLIP-qPCR analysis at the same D2 EpiLCs of Pspc1 WT/KO, Neat1 WT/KO, and Tet1 WT/KO (a genetic KO, see Dawlaty et al., 2011) genotypes. We first confirmed that EZH2 protein levels were not affected upon loss of Pspc1, Neat1, or Tet1 in ESCs and D2 EpiLCs (Figure S6D). PSPC1 mRNA and protein expression increased in D2 EpiLCs relative to ESCs (Figures 3F and S6D). PSPC1 also interacted with TET1 in D2 EpiLCs (Figure S6E). We then compared EZH2 binding to the transcripts of bivalent genes (e.g., Fgf5, Nefl, and Sall2) activated during the ESC-to-EpiLC transition (Figures 3E and 3F). We found enhanced EZH2 binding to these mRNA transcripts upon the loss of Pspc1 or Tet1 (Figure 6E), accompanied by decreased PRC2 chromatin binding at promoters (Figure 5H). However, EZH2 binding to these mRNA transcripts decreased upon the loss of Neat1 (Figure 6E). Next, we asked whether PSPC1 restricts EZH2 binding to bivalent transcripts is through PSPC1’s RNA-binding capacity. Using the Pspc1KO ESCs rescued with either PSPC1 WT or RRMmut protein (Figure S3C), we first verified that PSPC1 binding to Neat1 was significantly compromised in the PSPC1 RRMmut-rescued ESCs compared with the WT-rescued ESCs (Figure S6F). In D2 EpiLCs, we found that the WT-rescued but not the RRMmut-rescued Pspc1KO cells significantly reduced the heightened EZH2 binding to mRNA transcripts in Pspc1KO cells to a near-wild-type level (Figure S6G). We then addressed whether PSPC1 restricts EZH2 binding to bivalent gene transcripts through its RNA-binding capacity to Neat1 and/or bivalent gene transcripts. To this end, we performed PSPC1 CLIP-qPCR on Neat1 and bivalent gene transcripts in D2 EpiLCs. Interestingly, we found that PSPC1 RNA-binding capacity was specific only to Neat1 but not to the bivalent gene transcripts and was independent of TET1 (Figure 6F), which was distinct from the promiscuous RNA binding by PRC2 (Davidovich et al., 2013).
In sum, the enrichment of the Neat1 ChIRP intensity at the bivalent regions in both ESCs and EpiLCs (Figure 6D) and the preferential binding of PSPC1 to Neat1 help explain the requirement of Neat1 for the bivalent chromatin occupancy of PSPC1 (and its close partner TET1). As TET1 and PSPC1 inhibit, and Neat1 promotes (Figure 6E), the PRC2 binding to bivalent mRNA transcripts, these results support that PSPC1 and TET1 act through Neat1 to modulate PRC2 binding to bivalent gene transcripts and control stem cell bivalency.
DISCUSSION
Whereas a published study establishes a catalytic activity-dependent role of TET1 in demethylating bivalent promoters for the super-bivalency in formative pluripotency (Xiang et al., 2020), our study delineates a catalytic activity-independent role of TET1 in preventing hyper-activation of bivalent genes and thus preserving the bivalency in ESCs and during the ESC-to-EpiLC transition. Our data also support the PRC2 “eviction” models (Wang et al., 2017a, 2017b) and provide detailed mechanistic insight into the proposed repressive role of TET1 during bivalent gene activation (Koh et al., 2011; Wu et al., 2011). Our findings are in line with a recent study suggesting that TET1 regulates bivalent developmental genes independently of its catalytic activity (Chrysanthou et al., 2022). We thus establish a stem cell paradigm whereby TET1 and its close partner PSPC1 prevent transcriptional activation of bivalent genes in ESCs and fine-tune the bivalent gene transcription during the ESC-to-EpiLC transition by promoting PRC2 chromatin occupancy and H3K27me3 deposition at bivalent promoters and restricting the PRC2 binding to the bivalent gene transcripts, respectively, partly through Neat1-mediated interplay between PSPC1 and PRC2 (Figures 7A and 7D). In ESCs, while the loss of Pspc1 or Neat1 modifies the H3K27me3 distribution (Figure S5D), expression of bivalent genes is minimal (Figures 7B and 7C). Like Tet1KO ESCs (Dawlaty et al., 2011), Pspc1KO and Neat1KO ESCs maintain self-renewal and the expression of pluripotency-associated genes. However, in EpiLCs, upon the loss of Tet1 or Pspc1, Neat1 maintains its expression and positively mediates transcriptional activation of bivalent genes, likely through promoting PRC2 binding, directly or indirectly (see Limitations of the study), to the nascent mRNAs (Figures 7D and 7E). Our study thus provides mechanistic insights into how a dynamic balance between PRC2 chromatin occupancy and scanning of mRNAs is maintained during the ESC-to-EpiLC transition (indicated by the up/down dashed arrows of Figure 7D). Without Neat1 (i.e., Neat1KO), the balance of PRC2 chromatin occupancy and RNA binding may be altered in favor of the former, resulting in attenuated bivalent gene transcription (Figure 7F).
Figure 7. The working model of this study.
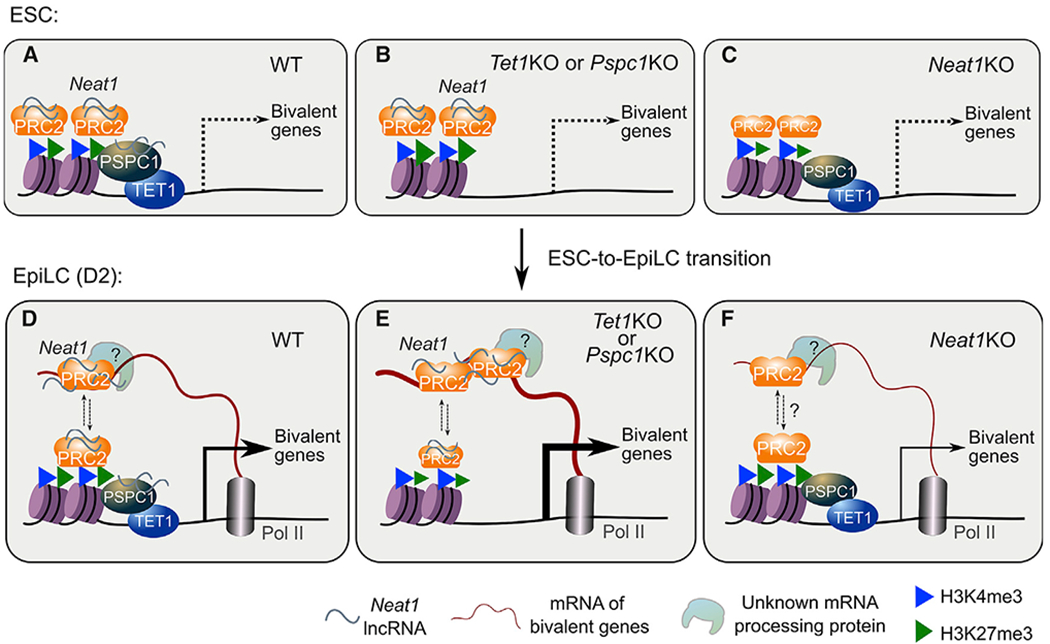
(A–C) In ESCs (WT), Neat1 (short isoform, Neat1_1) associates with the chromatin-bound proteins TET1, PSPC1, and PRC2 at bivalent gene promoters (A). Bivalent genes are minimally expressed in WT (A), Tet1KO, or Pspc1KO (B), or Neat1KO (C) ESCs. In Neat1KO ESCs and D2 EpiLCs(WT or KO), the chromatin-bound PSPC1 and TET1 decrease, denoted by smaller protein symbols. Bivalent genes are activated during pluripotent-state transition (accompanied by downregulation of Neat1_1, with no expression of Neat1_2 yet), and nascent mRNA acts as a decoy to evict PRC2 from chromatin.
(D–F) In EpiLCs(WT), a dynamic balance is maintained between PRC2 chromatin occupancy and RNA binding (shown in up/down arrows)to fine-tune the expression of bivalent genes (D). In Tet1KO or Pspc1KO (E) EpiLCs, more PRC2 proteins bind to mRNAs and are displaced or evicted from chromatin, inducing enhanced bivalent gene transcription. Without Neat1 (F), the balance between the chromatin- and mRNA-bound PRC2 may be disrupted (indicated by dashed lines and a question mark). PRC2-binding affinityto mRNAs (and possibly mRNA-processing-associated proteins) is compromised, which causes reduced bivalent gene activation. Of note, although PRC2 binds to both Neat1 and certain bivalent gene transcripts, Neat1 may promote PRC2 binding to nascent mRNA transcripts indirectly (D–F, e.g., through unknown mRNA-processing protein; see Limitations of the study).
While Neat1 function in modulating PRC2 chromatin occupancy was reported (Wang et al., 2019), its role in promoting PRC2 binding to bivalent gene transcripts when PSPC1 and/or TET1 are depleted (Figure 6E) is an unexpected finding. In recent years, phase separation in the regulation of gene transcription has become an area of intense research (Hnisz et al., 2017). RNA Pol II acts in gene transcription through phase separation (Lu et al., 2018), and Neat1 also scaffolds protein interactions of many RBPs that align to form paraspeckles by phase separation (Yamazaki et al., 2018). We recently revealed that PSPC1 promotes Pol II engagement and activity for the actively transcribed genes by enhancing the phase separation and subsequent phosphorylation and release of polymerase condensates (Shao et al., 2022). In our model, Neat1 may facilitate phase separation of other mRNA-processing proteins (i.e., ribonucleoprotein complex) for maintaining gene transcription and mRNA processing. This concept is supported by a recent proteomics study revealing that RNase treatment or Pol II inhibition reduces the chromatin fraction of RNA-processing proteins while increasing the chromatin fraction of transcription factors and chromatin modifiers (Skalska et al., 2021). The nascent mRNAs and other noncoding RNAs, including Neat1, may contribute to a dynamic matrix or phase-separated compartments that regulate chromatin states and gene transcription (Creamer et al., 2021; Skalska et al., 2021).
The lncRNA Neat1 has two isoforms. The long isoform Neat1_2 is essential for the assembly of paraspeckles (Jiang et al., 2017; Nakagawa et al., 2011). The short isoform Neat1_1, albeit also a paraspeckle component, plays various paraspeckle-independent roles (Fox et al., 2018; Li et al., 2017). Our Neat1 ChIRP-seq data (Figures 4F–4I) suggest that Neat1_1 may be necessary for proper activation of bivalent lineage genes by interaction with other RBPs (e.g., PSPC1 and EZH2) (Figures 6E and 6F) when their promoters are still bivalent (Figures 5F–5H). Accordingly, the “super-bivalency” at lineage-specific genes in formative pluripotency state may represent a few key molecular features, including the initiation of bivalent gene transcription, preservation of bivalent histone marks (H3K4me3 and H3K27me3), occupancy of chromatin-bound transcriptional co-factors (i.e., PSPC1 and TET1), and homeostasis of RNA-bound and chromatin-bound PRC2 (Figure 7D). PRC2 is known to bind to thousands of RNA transcripts with low specificity (Davidovich et al., 2013; Kaneko et al., 2013), including Neat1 and bivalent gene transcripts, through competition with various RBPs including PSPC1. In ESCs, since expression of bivalent gene transcripts is minimal, Pspc1KO may increase Neat1-mediated PRC2 recruitment and H3K27me3 (Wang et al., 2019). However, in EpiLCs, the change of transcription program (i.e., activation of bivalent gene transcripts) rebalances the PRC2 molecules that are available to chromatin, nascent transcripts, and/or Neat1. In Pspc1KO or RRMmut-rescued cells, the bivalent gene transcript-bound PRC2 increases (Figures 6E and S6G), likely through the abrogation of PSPC1-Neat1 interaction (Figures 6F and S6F) and thus more Neat1 available for PRC2 associations. Of note, the super-bivalency in formative pluripotency (D2 EpiLC) is likely a transient status because, during further differentiation (D4 EpiLCs or later), depletion of Neat1_1 and higher expression of the bivalent genes may eliminate (evict) PRC2 and repressive H3K27me3 on bivalent chromatin. During differentiation of human ESCs (hESCs), paraspeckles start to form with the expression of Neat1_2 (Modic et al., 2019). TDP43 post-transcriptionally regulates alternative polyadenylation (APA) of Neat1 to produce the long isoform Neat1_2 required for efficient early differentiation of hESCs (Grosch et al., 2020; Modic et al., 2019). Therefore, expression of Neat1_1, albeit lacking paraspeckle assembly, is conserved in both mouse and human pluripotency, akin to the conservation of stem cell bivalency in both mouse and human.
Limitations of the study
Our current study does not have direct evidence that the nascent mRNAs of bivalent genes are subjected to PRC2 dynamic binding during ESC-to-EpiLC transition, which requires CLIP-seq analysis of PRC2 in a combination of global run-on sequencing (GRO-seq) to measure the association of PRC2 with the nascent mRNAs. In addition, we acknowledge that we do not have data supporting that PRC2 directly interacts with both Neat1 and the nascent transcript. Although Neat1KO reduces the interactions between PRC2 and certain bivalent gene transcripts (Figure 6E), this could be an indirect effect resulting from alterations in other mRNA-processing proteins (indicated by in “?” in Figure 7D–F), given that Neat1KO does not lead to changes in the occupancy of PRC2 on chromatin in D2 EpiLCs (Figures 5F–5H). As discussed, we reported in another study that nascent RNAs could synergize with PSPC1 and promote Pol II activity by enhancing phase separation (Shao et al., 2022), although it remains to be determined whether Neat1 competes with or facilitates nascent RNAs in the polymerase condensates on bivalent genes.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jianlong Wang (jw3925@cumc.columbia.edu).
Materials availability
The Neat1KO and Tet1-degron ESC lines generated in this paper are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
The ChIP-seq, ChIRP-seq, and RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) with accession code: GSE182443. The TET1 affinity purification followed by mass spectrometry data have been deposited at the ProteomeXchange Consortium via the PRIDE partner repository with accession code: PXD033587. The deposited data are publicly available as of the date of publication. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| TET1 | Millipore | Cat. 09-872; RRID: AB_10806199 |
| TET1 | GeneTex | Cat. GTX125888; RRID: AB_11164485 |
| PSPC1 | Santa Cruz | Cat. sc-84577; RRID: AB_2171459 |
| PSPC1 | Bethyl | Cat. A303-206A; RRID: AB_10954256 |
| PSPC1 | Sigma | Cat. SAB4200503; RRID: N/A |
| EZH2 | Cell Signaling | Cat. 5246S; RRID: AB_10694683 |
| SUZ12 | Abcam | Cat. ab12073; RRID: AB_442939 |
| SUZ12 | Active Motif | Cat. 39357; RRID: AB_2614929 |
| V5 | Invitrogen | Cat. R960-25; RRID: AB_2556564 |
| Mouse IgG | Millipore | Cat. 12-371; RRID: AB_145840 |
| Rabbit IgG | Millipore | Cat. PP64; RRID: AB_97852 |
| SIN3A | Abcam | Cat. ab3479; RRID: AB_303839 |
| PELP1 | Bethyl | Cat. A300-180A; RRID: AB_242526 |
| TET2 | Abcam | Cat. ab124297; RRID: AB_2722695 |
| NONO | Bethyl | Cat. A300-587A; RRID: AB_495510 |
| SFPQ | Abcam | Cat. ab38148; RRID: AB_945424 |
| HA | Abcam | Cat. ab9110; RRID: AB_307019 |
| OCT4 | Santa Cruz | Cat. sc-5279; RRID: AB_628051 |
| NANOG | Bethyl | Cat. A300-397A; RRID: AB_386108 |
| ESRRB | R&D Systems | Cat. PP-H6707; RRID: AB_2100411 |
| ACTIN | Sigma | Cat. A5441; RRID: AB_476744 |
| GAPDH | ProteinTech | Cat. 10494-1-AP; RRID: AB_2263076 |
| Histone3 | Abcam | Cat. ab1791; RRID: AB_302613 |
| H3K4me3 | EpiCypher | Cat. 13-0041; RRID: N/A |
| H3K27me3 | Cell Signaling | Cat. 9733S; RRID: AB_2616029 |
| VCL | Abcam | Cat. ab129002; RRID: AB_11144129 |
| Streptavidin-HRP | GE Healthcare | Cat. RPN1231 V; RRID: N/A |
| Mouse IgG HRP | Cell Signaling | Cat. 7076S; RRID: AB_330924 |
| Rabbit IgG HRP | Jackson ImmunoRes | Cat. 715-175-151; RRID: AB_2340820 |
| Trueblot Mouse IgG HRP | Rockland | Cat. 18-8817-31; RRID: AB_2610850 |
| Trueblot Rabbit IgG HRP | Rockland | Cat. 18-8816-31; RRID: AB_2610847 |
| DNA 5mC | Cell Signaling | Cat. 28692; RRID: AB_2798962 |
| DNA 5hmC | Active Motif | Cat. 39769; RRID: AB_10013602 |
| Anti-dsDNA | Abcam | Cat. ab27156; RRID: AB_470907 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | GIBCO | Cat. 11965-092 |
| Heat inactivated FBS | GIBCO | Cat. 35-011-CV |
| Penicillin-Streptomycin | GIBCO | Cat. 15140-122 |
| L-Glutamine | GIBCO | Cat. 25030-081 |
| MEM NEAA | GIBCO | Cat. 11140-050 |
| 2-Mercaptoethanol | Sigma | Cat. M6250 |
| Puromycin | Sigma | Cat. P9620-10ML |
| Hygromycin | Omega | Cat. HG-80 |
| N2 | GIBCO | Cat. 17502-048 |
| B27 | GIBCO | Cat. 17504-044 |
| DMEM/F-12 | GIBCO | Cat. 11-330-032 |
| Neurobasal | GIBCO | Cat. 21-103-049 |
| LIF | Lab prep | N/A |
| GSK3i (CHIR99021) | Sigma | Cat. SML1046-25MG |
| MEKi (PD0325901) | Selleckchem | Cat. S1036 |
| Recombinant Fgf2 | R&D System | Cat. 233-FB |
| Recombinant Activin A | R&D System | Cat. 338-AC |
| 13C615N4 L-arginine | Cambridge Isotope | Cat. CNLM-539-H |
| 13C615N2 L-lysine | Cambridge Isotope | Cat. CNLM-291-H |
| 13C6 L-lysine | Cambridge Isotope | Cat. CLM-2247-H |
| dTAG-13 | Tocris | Cat. 6605 |
| Deposited data | ||
| PSPC1 ChIP-seq in ESC | This paper | NCBI GEO: GSE182443 |
| SUZ12 ChIP-seq in ESC | This paper | NCBI GEO: GSE182443 |
| SUZ12 ChIP-seq in EpiLC upon Pspc1 KO, Neat1 KO, and Tet1-degron treatments | This paper | NCBI GEO: GSE182443 |
| H3K27me3 ChIP-seq in ESC and EpiLC upon Pspc1 KO, Neat1 KO, and Tet1-degron treatments | This paper | NCBI GEO: GSE182443 |
| H3K4me3 ChIP-seq in ESC and EpiLC upon Pspc1 KO and Neat1 KO | This paper | NCBI GEO: GSE182443 |
| Neat1 ChIRP-seq in WT and Neat1 KO ESC and EpiLC | This paper | NCBI GEO: GSE182443 |
| Pspc1 WT/KO RNA-seq in ESC and EpiLC | This paper | NCBI GEO: GSE182443 |
| Neat1 WT/KO RNA-seq in ESC and EpiLC | This paper | NCBI GEO: GSE182443 |
| TET1 affinity purification followed by mass spectrometry data in ESC | This paper | ProteomeXchange PRIDE: PXD033587 |
| TET1 ChIP-seq in ESC | Wu et al., 2011 | NCBI GEO: GSE26833 |
| PSPC1 CLIP-seq in ESC | Guallar et al., 2018 | NCBI GEO: GSE103269 |
| H3K4me3 and H3K27ac in ESC | Hon et al., 2014 | NCBI GEO: GSE48519 |
| 5mC and 5hmC meDIP-seq in ESC | Xiong et al., 2016 | NCBI GEO: GSE57700 |
| Experimental models: Cell lines | ||
| Mouse ESC CCE | This paper | N/A |
| Pspc1 KO ESC | Guallar et al., 2018 | N/A |
| Pspc1 KO ESC rescued with WT or RRMmut PSPC1 protein | Guallar et al., 2018 | N/A |
| Tet1-degron ESC | This paper | N/A |
| Mouse ESC 46C | This paper | N/A |
| Neat1 KO ESC | This paper | N/A |
| Ezh2 KO ESC | Shen et al., 2008 | N/A |
| Mouse ESC V6.5 | Laboratory of R. Jaenisch | N/A |
| Tet1 KO ESCs | Laboratory of R. Jaenisch | N/A |
| Tet1/2/3 KO ESCs | Laboratory of R. Jaenisch | N/A |
| Dnmt1/3a/3b KO ESC | Laboratory of T. Chen | N/A |
| Nono KO ESC | Laboratory of F. Lan | N/A |
| Oligonucleotides | ||
| Oligonucleotides (see Table S4) | This paper | N/A |
| Software and algorithms | ||
| STAR | 2.7.6a | https://github.com/alexdobin/STAR |
| Cufflinks | 2.2.1 | http://cole-trapnell-lab.github.io/cufflinks/ |
| Bowtie2 | 2.3.5 | http://bowtie-bio.sourceforge.net/bowtie2/ |
| IGV | 2.10.2 | https://software.broadinstitute.org/software/igv |
| samtools | 1.10 | http://www.htslib.org/ |
| PICARD | 2.18.5 | https://broadinstitute.github.io/picard/ |
| HOMER | 4.11.1 | http://homer.ucsd.edu/homer/ |
| MACS2 | 2.2.7 | https://github.com/macs3-project/MACS |
| NGSplot | 2.61 | https://github.com/shenlab-sinai/ngsplot |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture and in vitro differentiation
If not specified, mouse embryonic stem cells (ESCs) were cultured on 0.1% gelatin-coated plates and in ES medium: DMEM medium supplemented with 15% fetal bovine serum (FBS), 1000 units/mL recombinant leukemia inhibitory factor (LIF), 0.1 mM 2-mercaptoethanol, 2 mM L-glutamine, 0.1 mM MEM non-essential amino acids (NEAA), 1% nucleoside mix (100X stock), and 50 U/mL Penicillin/Streptomycin. The Ezh2 KO ESCs (Shen et al., 2008) were cultured on 0.1% gelatin-coated plates and in naive culture condition (2iL) using serum-free N2B27 medium (DMEM/F12 and Neurobasal medium mixed at a ratio of 1:1, 1 × B27 supplement, 1 × N2 supplement, 2 mM L-glutamine, 0.1 mM 2-mercaptoethanol, and 50 U/mL Penicillin/Streptomycin) supplemented with Gsk3β inhibitor (CHIR99021, 3 μM), Mek inhibitor (PD0325901, 1 μM), and LIF (1000 units/mL).
For SILAC labeling, ESCs were cultured in either SILAC heavy or light medium: ES medium with complete supplements but deficient in both L-lysine and L-arginine, and then supplemented with L-lysine and L-arginine (SILAC light) or 13C615N4 L-arginine (Arg+10) and 13C615N2 L-lysine (Lys+8) or 13C6 L-lysine (Lys+6) (SILAC heavy) amino acids (Cambridge Isotope Laboratories).
For in vitro ESC-to-EpiLC differentiation, ESCs were seeded on fibronectin-coated (10 μg/mL/cm2) plates and in ES medium. On the next day, the medium was switched to formative culture condition using serum-free N2B27 medium supplemented with Fgf2 (12 ng/mL) and Activin A (20 ng/mL) (FA).
Neat1 knockout (KO) ESCs
CRISPR/Cas9-mediated Neat1KO was performed as described in (Yin et al., 2015). Briefly, two vectors (with the same pGL3-U6-sgRNA-PGK-puromycin backbone, Addgene #51133) containing two sgRNA sequences (Table S4) targeting a 6K bp region containing the short isoform of Neat1 (Neat1_1) were cotransfected with a Cas9-expressing vector (pST1374-N-NLS-flag-linker-Cas9, Addgene #44758) into WT 46C ESCs by lipofectamine 2000 (Invitrogen). Transfected cells were selected with puromycin and blasticidin for 8 days before clones were picked. Then, individual ESC clones were expanded and subjected to genomic DNA extraction and PCR for genotyping screening. The KO clones were further confirmed by RT-qPCR analysis of Neat1 expression.
Tet1-degron knock-in (KI) and protein degradation
The CRISPR/Cas9 system was used to engineer ESCs for protein degradation of TET1 genetically. The 5′- and −3″-homology arms of Tet1 were PCR amplified from genomic DNA. The P2A-2xHA-FKBP(F36 V) fragment for N-terminal insertion and the mCherry and BFP sequences were PCR amplified from Addgene plasmids #91792, #104370, #104371, respectively. Tet1 5′- and −3″-homology arms, FKBP, and mCherry or BFP sequences were assembled by Gibson Assembly 2× Master Mix (NEB, E2611S) to obtain 5′arm-FKBP-BPF-3′arm and 5′arm-FKBP-mCherry-3′arm doner vectors in pJET1.2 vector (Thermo Scientific). CRISPR gRNA was subcloned into the pSpCas9(BB)-2A-Puro (PX459) vector (gRNA sequence in Table S4). ESCs were transfected with the two donor vectors and CRISPR vectors using Lipofectamine 2000 (Invitrogen). After two days of puromycin selection, double-positive cells were sorted out for mCherry and BFP and seeded on a 96-well plate with single-cell per well using the BD Influx Cell Sorter. Cells were expanded and genotyped by PCR, and protein degradation was confirmed by Western blot analysis. Clones with a homozygous knock-in tag were further expanded and used for experiments.
The Tet1-degron ESCs were treated with either DMSO control or dTAG13 (500 nM in DMSO, Tocris, 6605) for rapid degradation of TET1 protein. ESCs were treated with dTAG13 for 2 days before differentiation, and then cells were treated with dTAG13 during the ESC-to-EpiLC differentiation. In the control group, cells were treated with DMSO in ESCs and during the ESC-to-EpiLC differentiation.
METHOD DETAILS
Affinity purification followed by mass spectrometry (AP-MS) analysis
We employed a previously validated ESC clone with the ectopic expression of the 3xFLAG tagged mouse Tet1 (FL-Tet1) gene (Ding et al., 2015). Before the AP-MS experiment, the empty vector (EV)- and FL-Tet1-transfected ESCs were cultured in both SILAC heavy and light medium for 2 weeks with reciprocal labeling: Replicate#1, light of FL-Tet1 versus heavy of EV (Lys+6); Replicate#2, light of EV versus heavy of FL-Tet1 (Lys+8, Arg+10). AP-MS was performed using our well-established protocols (Ding et al., 2015; Guallar et al., 2018; Huang et al., 2021). Briefly, the cell pellets were resuspended in ice-cold hypotonic buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2 mM PMSF, and protease inhibitor cocktail (PIC, Sigma, P8340)) and incubated for 10 min on ice. The sample was centrifuged at 3,000 ×g for 5 min at 4°C, and the pellet containing nuclei was washed by resuspending with ice-cold buffer A and centrifuging at 10,000 × g for 20 min at 4°C. Then, nuclei were resuspended with ice-cold nuclear extract buffer C (20 mM HEPES, pH 7.9, 20% glycerol (v/v), 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, and PIC) and incubated at 4°C for 30 min with continuous mixing. Insoluble materials were pelleted by centrifugation at 25,000 × g for 20 min at 4°C. The supernatant was collected as nuclear extract (NE) and dialyzed against buffer D (20 mM HEPES, pH 7.9, 20% glycerol (v/v), 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF) for 3 h at 4°C. Then, 0.1 mL of Protein G agarose (Roche Diagnostic) equilibrated in buffer D containing 0.02% NP40 (buffer D-NP) was added to nuclear extracts in 15 mL tubes, in the presence of Benzonase (25 U/mL, Millipore 70664), and incubated/pre-cleared for 1 h at 4°C with continuous mixing. Precleared NE samples were incubated with pre-equilibrated anti-FLAG M2 affinity gel (Sigma, F2426) for 4 h at 4°C with continuous mixing. Five washes were performed with buffer D-NP. Bound material was eluted by incubation with buffer D-NP supplemented with 0.5 mg/mL 3xFLAG peptides (Sigma, F4799) for 2 h at 4°C with continuous mixing. The eluted proteins were concentrated with Amicon Ultra Centrifugal Filters (Millipore, UFC500396), boiled 5 min in Laemmli buffer, and fractionated on a 10% SDS-PAGE gel. The gel lanes were cut horizontally into 5~7 pieces, and each was subjected to LC-MS/MS analysis (Huang et al., 2021).
MS data were processed by Thermo Proteome Discoverer software with SEQUEST engine against mouse International Protein Index (IPI v3.68) protein sequence database. Carbamidomethylation (CAM) was set as the fixed modification, and methionine oxidation was set as the variable modification. Outputs of protein identification from Proteome Discoverer were imported into a local Microsoft Access database. Common contamination proteins (trypsin, keratins) were removed, and protein Heavy/Light quantification ratios were obtained.
Co-immunoprecipitation (co-IP)
Co-IP in regular (nucleosome-free) conditions was performed as previously described (Ding et al., 2015). The nuclei were purified with buffer A followed the AP-MS protocol. Then nuclei were resuspended with ice-cold lysis buffer (50 mM HEPES, pH 7.9, 250 mM NaCl, 0.1% NP-40, 0.2 mM EDTA, 0.2 mM PMSF, and PIC) and incubated at 4°C for 30 min with continuous mixing. About 2% of input was saved, then NE was diluted with 40% volume (v/v = 5:2) of dilution buffer (20 mM HEPES, pH 7.9, 20% glycerol (v/v), 0.05% NP-40, 0.2 mM EDTA, 0.2 mM PMSF, and PIC) as the co-IP buffer with the NaCl concentration of 180 mM. For antibody IP, the antibody and the same amount of mouse or rabbit IgG as control were added to the co-IP buffer, incubated with protein lysates overnight at 4°C with continuous mixing. Then, protein lysates were incubated with protein G-Agarose beads (Roche, 11243233001) for 2 h at 4°C with continuous mixing. For FLAG-IP, NE was incubated with anti-FLAG M2 affinity gel (Sigma, F2426) overnight at 4°C with continuous mixing. Beads were washed 4X with co-IP buffer (lysis buffer/dilution buffer = 5:2, v/v). For RNase A treatment, the beads were split during the first wash and incubated with or without RNase A (200 μg/mL, Sigma, R6148) at 37°C for 15 min. Proteins were eluted from the beads by boiling in 1X SDS Laemmli loading buffer, followed by SDS-PAGE and Western blot analysis.
Co-IP in nucleosome-containing conditions was performed following a published protocol (Neri et al., 2013). Briefly, cell pellets were resuspended in isotonic buffer (20 mM HEPES, pH 7.5, 100 mM NaCl, 250 mM Sucrose, 5 mM MgCl2, 5 μM ZnCl2, and PIC), incubated on ice for 5 min, and spun down 500 g for 5 min at 4°C. Then pellets were resuspended in isotonic buffer (no PIC) supplemented with 1% NP-40), vortexed for 10 s at the highest setting, incubated on ice for 5 min, and spun down 1000 g for 5 min at 4°C. The pellets (nuclei) were resuspended in 200 μL digestion buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 250 mM Sucrose, 0.5 mM MgCl2, 5 mM CaCl2, 5 μM ZnCl2, no PIC) and 1 μL of micrococcal nuclease (MNase, NEB, M0247S), incubated at 37°C water bath for 10 min. Then the MNase digestion was immediately stopped by adding 20 μL 0.5 M EDTA, and nuclei were spun down 13,000 g for 1 min at 4°C. The digested nuclei were resuspended in digestion buffer (with PIC), subjected to sonication with Bioruptor Plus, set 30 s ON, 30 s OFF, 5 cycles to break nuclei, and spun down 13,000 g for 5 min at 4°C. Protein supernatants were subjected to antibody incubation, washing with digestion buffer, protein elution, and SDS-PAGE, like the regular co-IP protocol.
The primary antibodies used for co-IP were: TET1 (Millipore, 09-872 and GeneTex, GTX125888), PSPC1 (Santa Cruz, sc-84577 and Bethyl, A303-206A), SUZ12 (Abcam, ab12073), EZH2 (Cell Signaling, 5246S), V5 (Invitrogen, R960-25), mouse IgG (Millipore, 12-371), and rabbit IgG (Millipore, PP64).
Subcellular fractionation assay
The subcellular fractions of ESCs were extracted using the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo, #78840). Briefly, about 5 × 106 cells were used, and each subcellular fraction was collected following the standard protocol. Protein loadings were balanced according to the protein concentrations in the cytoplasmic fraction before Western blot analysis.
Gel filtration assay
Size exclusion chromatography (gel filtration assay) was performed as previously described (Ding et al., 2015). Briefly, nuclear extracts (10~20 mg) of ESCs were applied to a gel filtration column (S400 HiPrep 16/60 Sephacryl, Amersham Biosciences), samples were eluted at 1 mL/min and continuously monitored with an online detector at a wavelength of 280 nm. Fractions were collected, concentrated, and subjected to Western blot analysis with indicated antibodies.
Domain mapping
The FLAG-tagged Tet1 full-length (FL) sequence and truncated variants were cloned in the PiggyBac expression vectors. The Pspc1 full-length sequence and truncated variants were PCR amplified and subcloned into the V5-tagged PiggyBac expression vectors. The TET1 and PSPC1 PiggyBac expression vectors and control empty vectors (EV) were transfected into ESCs with Lipofectamine 2000 Transfection Reagent (Invitrogen, 11668019) following the standard protocol. After drug selection, ESCs were expanded for co-IP. FLAG-IP (for TET1 FL and truncated variants) and V5-IP (for PSPC1 FL and truncated variants) were performed, followed by Western blot analysis of PSPC1 and TET1, respectively.
Western blot analysis
Western blot analysis was performed as previously described (Huang et al., 2017). Total proteins were extracted by RIPA buffer. Protein concentrations were measured by Bradford assay (Pierce, 23236), balanced, and subjected to SDS-PAGE analysis. The following primary antibodies were used: PSPC1 (Bethyl, A303-206A and Sigma, SAB4200503), TET1 (Millipore, 09-872 and GeneTex, GTX125888), SIN3A (Abcam, ab3479), PELP1 (Bethyl, A300-180A), TET2 (Abcam, ab124297), V5 (Invitrogen, R960-25), NONO (Bethyl, A300-587A), SFPQ (Abcam, ab38148), HA (Abcam, ab9110), OCT4 (Santa Cruz, sc-5279), ESRRB (R&D, PP-H6707), NANOG (Bethyl, A300-397A), SUZ12 (Abcam, ab12073), EZH2 (Cell Signaling, 5246S), ACTIN (1:5000, Sigma, A5441), GAPDH (ProteinTech, 10494-1-AP), Histone 3 (H3, Abcam, ab1791), and Vinculin (VCL, Abcam, ab129002).
Immunofluorescence
Mouse embryonic fibroblasts (MEFs) and ESCs were grown on 24-well plates coated with 0.1% gelatin (w/v). After fixation with 4% paraformaldehyde (w/v) for 15 min, cells were permeabilized with 0.25% Triton X-100 (v/v) in PBS for 5 min and incubated with 10% BSA for 30 min at 37°C. For immunostaining, cells were incubated overnight at 4°C with PSPC1 antibody (Santa Cruz, sc-84577) in PBS with 3% BSA (w/v). The following day cells were incubated with fluorophore-labeled secondary antibodies for 1 h at RT. Cells were imaged with a Leica DMI 6000 inverted microscope.
Dot blot analysis
The genomic DNA dot-blot analysis of 5mC and 5hmC was performed following the DNA Dot Blot Protocol (Cell Signaling, #28692) with modifications. Briefly, genomic DNA of ESCs was extracted using Quick-DNA Miniprep Plus Kit (Zymo Research, D4068), and DNA concentration was measured by NanoDrop. Next, the same amount of DNA was denatured with 10X DNA denaturing buffer (1 M NaOH and 0.1 M EDTA) and incubated at 95°C for 10 min, which was then immediately mixed with an equal volume of 20X SSC buffer, pH 7.0 (Invitrogen, 15557044) and chilled on ice. The DNA samples were diluted with a pre-determined amount and loaded on the positive-charged Nelyon membrane (GE Amersham, RPN2020B) using a vacuum chamber (Minifold, SRC-96). The membrane was dried, auto-crosslinked with 1200 × 100 μJ/cm2, and blocked with 5% milk/TBST for 1 h. Next, the membrane was incubated with 5mC (Cell Signaling, 28692) or 5hmC (Active Motif, 39769) antibodies, the same as the Western blot analysis. Then, the membrane was stripped with the stripping buffer (Thermo Scientific, 21059) and reblotted with the dsDNA (Abcam, ab27156) antibody as the loading control.
Biotinylated RNA synthesis, dot blot, and pull-down assay
Neat1-sense (Neat1-S) and antisense (Neat1-AS) DNAs were amplified with primers containing the T7 promoter sequence at the 5′end (Table S4) from a pJET1.2 cloning vector (Thermo Scientific, K1232) containing the Neat1_1 cDNA sequence. Linearized DNA was biotin-labeled and in vitro transcribed using the Biotin RNA Labeling Mix (Roche, 11685597910) and MEGAscript T7 Transcription Kit (Invitrogen, AM1333). Synthesized RNA was purified with the RNA Clean & Concentrator Kit (Zymo Research, R1015). The ESC total RNA and biotinylated RNA were loaded on the positive-charged Nelyon membrane (GE Amersham, RPN2020B), auto-crosslinked with 1200 × 100 μJ/cm2, and blocked with 5% milk/TBST for 1 h. Then the membrane was washed with TBST and incubated in TBST containing HRP-Conjugated Streptavidin (GE Healthcare, RPN1231 V) at room temperature (RT) for 2 h. The rest steps were the same as the Western blot analysis.
The biotinylated RNA pull-down assay was performed as previously described (Rinn et al., 2007). The ESC cell pellets were washed 2X with buffer A as in the co-IP protocol. Then nuclei were resuspended with RIP lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 1 mM EDTA, with PMSF, PIC, and RNase inhibitor) and incubated at 4°C for 30 min with continuous mixing. After centrifuge, nuclear extracts were supplied with tRNA (0.1 μg/μL, Roche, 10109541001) and incubated with 4 μg Neat1-S, Neat1-AS RNAs, or the beads-only fraction for 1 h at 4°C. Then 40 uL of Streptavidin M280 dynabeads (Invitrogen, 11205D) were added to each binding fraction and further incubated for 1 h at 4°C. Beads were washed 5X with the RIP buffer and boiled, followed by SDS-PAGE analysis.
Genomic DNA 5mC and 5hmC quantification by mass spectrometry
The UHPLC-MS/MS analysis for 5mC and 5hmC quantification was performed as previously described (Lai et al., 2018) on an Agilent 1290 Infinity II ultrahigh performance LC system coupled with an Agilent 6470 triple quadrupole mass spectrometer equipped with a jet stream electrospray ionization source (Santa Clara, CA). The mass spectrometer was operated under positive ionization using multiple reactions monitoring (MRM) mode. The selective MRM transitions were monitored as follows: m/z 242 → 83 for 5mC and m/z 258 → 142 for 5hmC. The frequencies of 5mdC and 5hmC over total deoxycytidine (dC) were calibrated by their corresponding stable isotope-labeled internal standards.
RT-qPCR
Total RNA was extracted using the GeneJet RNA Purification Kit (Thermo Scientific, K0732). Reverse transcription was performed, and cDNA was generated using the qScript kit (Quanta, 95048). Relative expression levels were determined using a QuantStudio 5 Real-Time PCR System (Applied Biosystems). Gene expression levels were normalized to Gapdh. Primers for RT-qPCR are listed in Table S4.
Chromatin immunoprecipitation (ChIP) and sequencing
ChIP assays were performed as previously described (Huang et al., 2017). Briefly, cell pellets were crosslinked with 1% (w/v) formaldehyde for 10 min at RT, followed by the addition of 125 mM glycine to stop the reaction. Next, chromatin extracts were sonicated into 200–500 bp with Bioruptor Plus (settings of 30 s ON, 30 s OFF, 30 cycles) or with Bioruptor Pico (settings of 30 s ON, 30 s OFF, 15 cycles). Immunoprecipitation was performed with the following primary antibodies: PSPC1 (Santa Cruz, sc-84577 and Bethyl, A303-206A), SUZ12 (Active Motif, 39357), H3K4me3 (EpiCypher, 13-0041), H3K27me3 (Cell Signaling, 9733S), TET1 (GenTex, GTX125888), or rabbit IgG (Millipore, PP64) overnight at 4°C with continuous mixing, followed by incubation with protein G dynaberads (Invitrogen, 10004D) for another 2 h at 4°C. The immunoprecipitated DNA was washed with ChIP RIPA buffer and purified with ChIP DNA Clean & Concentrator columns (Zymo Research, D5205). qPCR was performed with Roche SYBR Green reagents and a LightCycler480 (Roche) machine. Percentages of input recovery were calculated. The ChIP-qPCR primers are listed in Table S4.
For ChIP-seq, 10% of sonicated genomic DNA was used as ChIP input. Libraries were prepared using the NEBNext Ultra II DNA library prep kit and index primers sets (NEB, 7645S, E7335S) following the standard protocol. Sequencing was performed with the Illumina HiSeq 4000 Sequencer according to the manufacturer’s protocol. Libraries were sequenced as 150-bp paired-end reads.
Chromatin Isolation by RNA purification (ChIRP) and sequencing
ChIRP assays were performed following an established protocol (Chu et al., 2011) with modifications. Briefly, 32 anti-sense oligo probes covering the whole Neat1_1 lncRNA (3.2K bp, 1 probe/100 bp of RNA length) were designed using singlemoleculefish.com. The probes were separated into “odd” and “even” pools before the experiment. Cell pellets were harvested and crosslinked with 1% of glutaraldehyde in PBS for 10 min at RT, followed by the addition of 125 mM glycine to stop the reaction. Next, chromatin extracts were sonicated into 100–500 bp with Bioruptor Pico, set 30 s ON, 30 s OFF, 45 cycles. Sonication efficiency was checked by running a 1.5% agarose gel. The lysates after sonication were diluted with 2X volume of hybridization buffer and incubated with odd and even probe pools at 37°C hybridization oven for 4 h with rotation, followed by incubation of streptavidin C1 dynabeads (Invitrogen, 65001) for another 30 min. The dynabeads were washed 5X with wash buffer, and ChIRP RNA was purified from 10% beads to examine the Neat1 RNA enrichment. The ChIRP DNA was eluted from the remaining beads with elution buffer containing RNase A and RNase H and further treated with proteinase K. DNA was purified with Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v, Invitrogen, 15593031) and resuspended in TE buffer. ChIRP-seq libraries were prepared using the NEBNext Ultra II DNA library prep kit and index primers sets (NEB, 7645S, E7335S) following the standard protocol. Sequencing was performed with the Illumina HiSeq 4000 Sequencer according to the manufacturer’s protocol. Libraries were sequenced as 150-bp paired-end reads.
Crosslinking immunoprecipitation (CLIP) qPCR
UV crosslinking and immunoprecipitation (CLIP) were performed according to the eCLIP-seq protocol (Van Nostrand et al., 2016) with modifications. Briefly, cells in culture were washed with ice-cold PBS and crosslinked in PBS with UV type C (254 nm) at 400 mJ/cm2 on ice. Next, cells were scraped, pelleted, and lysed in CLIP lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with proteinase and RNase inhibitors, and incubated on ice for 1 h. The lysate was briefly sonicated with Bioruptor Plus, set 30 s ON, 30 s OFF, 5 cycles to break DNA. Next, Turbo DNase (2 U/μL, 1:500, Invitrogen, AM2238) was added, and the lysate was incubated in a 37°C water bath for 15 min, followed by centrifuge 15,000 g for 15 min at 4°C. Primary antibodies PSPC1 (Bethyl, A303-206A) and EZH2 (Cell Signaling, 5246S) or rabbit IgG (Millipore, PP64) were incubated with Protein-G dynabeads (Invitrogen) for 1 h at RT. Then the lysate and the beads were mixed overnight at 4°C with rotation. The next day, the beads were washed with wash buffer (low salt, 20 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 0.2% Tween-20) and high salt wash buffer (50 mM Tris-HCl, pH 7.4, 1 M NaCl), and digested with Proteins K to elute RNA. The input and CLIP RNAs were purified with the RNA Clean & Concentrator-5 kit (Zymo, R1015) followed by RT-qPCR analysis. Percentages of input recovery were calculated. CLIP-qPCR primer sequences are listed in Table S4.
Gene ontology (GO) analysis
Gene ontology (GO) analyses were performed using the DAVID gene ontology functional annotation tool (https://david.ncifcrf.gov/tools.jsp) with all NCBI Mus musculus genes as a reference list.
QUANTIFICATION AND STATISTICAL ANALYSIS
ChIP-seq and ChIRP-seq data processing
ChIP-seq data of histone marks H3K4me3 and H3K27ac in ESCs were downloaded from GSE48519, data of H3K27me3 in ESCs were downloaded from GSE89211 (Cruz-Molina et al., 2017), and data of TET1 in ESCs were downloaded from GSE26832. All ChIP-seq and ChIRP-seq reads were pre-processed by trim_galore (v0.6.3) and aligned to the mm9 mouse genome using the bowtie2 (v2.3.4) program, and the parameters were “-X 1000 –no-mixed –no-discordant”. The aligned reads were exported (-F 0x04 -f 0x02) and sorted with samtools. Duplicates were removed with MarkDuplicates function in the PICARD (v2.14.0) package. The aligned ChIP-seq and ChIRP-seq bam files of biological replicates were combined. All bam files were converted to a binary tiled file (tdf) and visualized using IGV (v2.7.2) software.
All ChIP-seq and ChIRP-seq peaks were determined by the MACS2 program (v.2.0.10). PSPC1 ChIP peaks in WT cells were called using the Pspc1KO ChIP-seq as the control data, and other ChIP peaks were called using the input ChIP-seq as the control data. Neat1 ChIRP peaks in WT cells were called using the Neat1KO ChIRP-seq as the control data. Broad peaks were called for PSPC1, SUZ12, and histone marks data, narrow peaks were called for TET1 and Neat1 data, and all other parameters were the default settings. All peaks were annotated using the annotatePeaks module in the HOMER program (v4.11) against the mm9 genome. A target gene of a called peak was defined as the nearest gene’s transcription start site (TSS) with a distance to TSS less than 5 kb. Heatmaps and mean intensity curves of ChIP-seq data at specific genomic regions were plotted by the NGSplot program (v2.61) centered by the middle point “(start+end)/2” of each region.
ChIP-seq correlation analysis of PSPC1 and other factors was performed with an in-house Python program as previously described (Ding et al., 2015). A phi correlation coefficient was used to calculate the correlation between the ChIP peaks of every two ChIP-seq data. Heatmap of correlations was shown with the Java TreeView (v1.1.6) program.
5mC and 5hmC DNA immunoprecipitation (DIP) data analysis
5mC and 5hmC DIP-seq data in ESCs were downloaded from GSE57700. Reads were aligned to the mouse genome mm9 using the bowtie (v1.0.0) program, with parameters -m 1 -v 2 –best –strata. The duplicated reads of the aligned data were removed, then filtered reads were sorted with samtools (v0.1.19). The reads per million (RPM) values of 5mC and 5hmC DIP-seq at each TET1 ChIP-seq peak region were calculated with the NGSplot program (v2.61)and shown in Boxplots using R. p value was calculated from two-sided Mann-Whitney test.
RNA-seq and data analysis
100 ng total RNA was processed for RNA-seq library construction using the Ovation Mouse RNA-seq kit (NuGEN, #0348–32) following the manufacturer’s protocol. Massively parallel sequencing was performed on an Illumina HiSeq 4000 Sequencing System. Libraries were sequenced as 150-bp paired-end reads. For RNA-seq data processing, reads were aligned to the mouse genome mm9 using STAR (v2.7.6a) with the default settings. Transcript assembly and differential expression analyses were performed using Cufflinks (v2.2.1). Assembly of novel transcripts was not allowed (-G). Other parameters of Cufflinks were the default setting. The summed FPKM (fragments per kilobase per million mapped reads) of transcripts sharing each gene_id was calculated and exported by the Cuffdiff program. In the gene expression matrix, a value of FPKM+1 was applied to minimize the effect of low-expression genes. p-values were calculated using a T-test. Differentially expressed genes (DEGs) were determined by two-sided T-test p-value<0.05 and fold-change>1.5. Boxplots for expression were generated using R. P-value was calculated from the two-sided Mann-Whitney test.
PCA-analysis was performed for RNA-seq data from different batches. Batch effects were adjusted by ComBat function implemented in the sva Bioconductor package (v.3.18.0). The expression data matrix was imported by Cluster 3.0 software for PCA analysis. PC values were visualized with the plot3d function in the rgl package using R (v4.1.0) scripts.
Statistical analysis
If not specified, qPCR analysis was performed in technical triplicates, and the error bars indicate standard deviation of the mean. p-values were calculated using a two-sided T-test in the GraphPad Prism software (v9.2.0). RT-qPCR analyses in Figures 3F, 4E, and S5B were performed in two independent KO clones. CLIP-qPCR analyses in Figures 6C, 6E, 6F, S6C, S6F, and S6G were repeated in biological duplicates.
The boxplots in Figures 4H, S2B, and S2E present the 25th, median, and 75th quartiles, and the whiskers extend 1.5 of interquartile ranges, and the p-value was calculated from the two-sided Mann-Whitney test. In the scatter plots of Figures 4C and S4E, P-value was calculated using the Fisher-exact test based on the number of DEGs in each category (Pspc1KO Up/Down vs. Neat1KO Up/Down). In the scatter plot of Figure S4D, Pearson’s product-moment correlation coefficient (r) and P-value of correlation are indicated in each plot. The statistical analysis was performed with R (v4.1.0) scripts on the R-Studio platform (v1.4.1). The statistical details of experiment are indicated in the figure legend.
Supplementary Material
Highlights.
The TET1 interactome identifies PSPC1 as a partner of TET1 in ESCs
PSPC1 interacts with TET1 and PRC2 for bivalency control in formative pluripotency
TET1 and PSPC1 repress bivalent genes by promoting PRC2 chromatin occupancy
Neat1 facilitates bivalent gene activation by promoting PRC2 binding to their mRNAs
ACKNOWLEDGMENTS
We thank Dr. Wei Xie for providing the Tet1 vectors for domain mapping, Dr. Fei Lan for providing the NonoKO ESCs, Dr. Taiping Chen for providing the Dnmt1/3a/3bTKO ESCs, Dr. Rudolf Jaenisch for providing the Tet1KO ESCs, and Dr. Francesco Neri for discussion on the TET1 co-IP protocol. Research in the Shen Laboratory was supported in part by the National Natural Science Foundation of China (31829003). This work in the Wang laboratory is funded by grants from the National Institutes of Health (R01GM129157, R01HD095938, R01HD097268, and R01HL146664) and by contracts from New York State Stem Cell Science (NYSTEM#C35583GG and C35584GG).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110928.
REFERENCES
- Beltran M, Yates CM, Skalska L, Dawson M, Reis FP, Viiri K, Fisher CL, Sibley CR, Foster BM, Bartke T, et al. (2016). The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 26, 896–907. 10.1101/gr.197632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bonetti A, Agostini F, Suzuki AM, Hashimoto K, Pascarella G, Gimenez J, Roos L, Nash AJ, Ghilotti M, Cameron CJF, et al. (2020). RADICL-seq identifies general and cell type-specific principles of genome-wide RNA-chromatin interactions. Nat. Commun 11, 1018. 10.1038/s41467-020-14337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353. 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, and Wysocka J (2014). Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14, 838–853. 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, and Carmichael GG (2009). Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell 35, 467–478. 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysanthou S, Tang Q, Lee J, Taylor SJ, Zhao Y, Steidl U, Zheng D, and Dawlaty MM (2022). The DNA dioxygenase Tet1 regulates H3K27 modification and embryonic stem cell biology independent of its catalytic activity. Nucleic Acids Res. 50, 3169–3189. 10.1093/nar/gkac089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, and Chang HY (2011). Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678. 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Hernandez AJ, Sarma K, and Lee JT (2014). Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell 55, 171–185. 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer KM, Kolpa HJ, and Lawrence JB (2021). Nascent RNA scaffolds contribute to chromosome territory architecture and counter chromatin compaction. Mol. Cell 81, 3509–3525.e5. 10.1016/j.molcel.2021.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Molina S, Respuela P, Tebartz C, Kolovos P, Nikolic M, Fueyo R, van Ijcken WFJ, Grosveld F, Frommolt P, Bazzi H, and Rada-Iglesias A (2017). PRC2 facilitates the regulatory topology required for poised enhancer function during pluripotent stem cell differentiation. Cell Stem Cell 20, 689–705.e9. 10.1016/j.stem.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Davidovich C, and Cech TR (2015). The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA 21, 2007–2022. 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, and Cech TR (2015). Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell 57, 552–558. 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, and Cech TR (2013). Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol 20, 1250–1257. 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, and Jaenisch R (2011). Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175. 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. (2013). TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655. 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Huang X, Shao N, Zhou H, Lee DF, Faiola F, Fidalgo M, Guallar D, Saunders A, Shliaha PV, et al. (2015). Tex10 coordinates epigenetic control of super-enhancer activity in pluripotency and reprogramming. Cell Stem Cell 16, 653–668. 10.1016/j.stem.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M, Huang X, Guallar D, Sanchez-Priego C, Valdes VJ, Saunders A, Ding J, Wu WS, Clavel C, and Wang J (2016). Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell 19, 355–369. 10.1016/j.stem.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, and Muller S (2011). The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 30, 1067–1078. 10.1038/emboj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Nakagawa S, Hirose T, and Bond CS (2018). Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci 43, 124–135. 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Grosch M, Ittermann S, Rusha E, Greisle T, Ori C, Truong DJJ, O’Neill AC, Pertek A, Westmeyer GG, and Drukker M. (2020). Nucleus size and DNA accessibility are linked to the regulation of paraspeckle formation in cellular differentiation. BMC Biol. 18, 42. 10.1186/s12915-020-00770-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guallar D, Bi X, Pardavila JA, Huang X, Saenz C, Shi X, Zhou H, Faiola F, Ding J, Haruehanroengra P, et al. (2018). RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet 50, 443–451. 10.1038/s41588-018-0060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, and Saitou M (2011). Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532. 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- He C, Sidoli S, Warneford-Thomson R, Tatomer DC, Wilusz JE, Garcia BA, and Bonasio R. (2016). High-resolution mapping of RNA-binding regions in the nuclear Proteome of embryonic stem cells. Mol. Cell 64, 416–430. 10.1016/j.molcel.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, and Sharp PA (2017). A phase separation model for transcriptional control. Cell 169, 13–23. 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao SQ, Wang BA, et al. (2014). 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol. Cell 56, 286–297. 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Balmer S, Yang F, Fidalgo M, Li D, Guallar D, Hadjantonakis AK, and Wang J (2017). Zfp281 is essential for mouse epiblast maturation through transcriptional and epigenetic control of Nodal signaling. Elife 6, e33333. 10.7554/elife.33333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Park K.m., Gontarz P, Zhang B, Pan J, McKenzie Z, Fischer LA, Dong C, Dietmann S, Xing X, et al. (2021). OCT4 cooperates with distinctATP-dependent chromatin remodelers in naïve and primed pluripotent states in human. Nat. Commun 12, 5123. 10.1038/s41467-021-25107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe M, Toya H, Mito M, Chiba T, Asahara H, Hirose T, and Nakagawa S (2020). Forced isoform switching of Neat1_1 to Neat1_2 leads to the loss of Neat1_1 and the hyperformation of paraspeckles but does not affect the development and growth of mice. RNA 26, 251–264. 10.1261/rna.072587.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Shao C, Wu QJ, Chen G, Zhou J, Yang B, Li H, Gou LT, Zhang Y, Wang Y, et al. (2017). NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol 24, 816–824. 10.1038/nsmb.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, and Reinberg D (2014). Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 53, 290–300. 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, and Bonasio R (2013). PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol 20, 1258–1264. 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoueiry R, Sohni A, Thienpont B, Luo X, Velde JV, Bartoccetti M, Boeckx B, Zwijsen A, Rao A, Lambrechts D, and Koh KP (2017). Lineage-specific functions of TET1 in the postimplantation mouse embryo. Nat. Genet 49, 1061–1072. 10.1038/ng.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Barber M, Mansfield W, Cui Y, Spindlow D, Stirparo GG, Dietmann S, Nichols J, and Smith A (2021). Capture of mouse and human stem cells with features of formative pluripotency. Cell Stem Cell 28, 2180–2471.e458. 10.1016/j.stem.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott GJ, Bond CS, and Fox AH (2016). The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res. 44, 3989–4004. 10.1093/nar/gkw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. (2011). Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213. 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, and Zhang Y (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502,472–479. 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W, Lyu C, and Wang H (2018). Vertical ultrafiltration-facilitated DNA digestion for rapid and sensitive UHPLC-MS/MS detection ofDNAmodifications. Anal. Chem 90, 6859–6866. 10.1021/acs.analchem.8b01041. [DOI] [PubMed] [Google Scholar]
- Li R, Harvey AR, Hodgetts SI, and Fox AH (2017). Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA 23, 872–881. 10.1261/rna.059477.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Karwacki-Neisius V, Ma C, Tan L, Shi Y, Wu F, and Shi YG (2020). Nono deficiency compromises TET1 chromatin association and impedes neuronal differentiation of mouse embryonic stem cells. Nucleic Acids Res. 48, 4827–4838. 10.1093/nar/gkaa213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Hwang T, Gooding AR, Goodrich KJ, Rinn JL, and Cech TR (2020). RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat. Genet 52, 931–938. 10.1038/s41588-020-0662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Liu Y, Jiang L, Yamaguchi S, and Zhang Y (2014). Role of Tet proteins in enhancer activity and telomere elongation. Genes Develop. 28, 2103–2119. 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, Darzacq X, and Zhou Q(2018). Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323. 10.1038/s41586-018-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Karwacki-Neisius V, Tang H, Li W, Shi Z, Hu H,Xu W, Wang Z, Kong L, Lv R, et al. (2016). Nono, a bivalent domain factor, regulates Erk signaling and mouse embryonic stem cell pluripotency. Cell Rep. 17, 997–1007. 10.1016/j.celrep.2016.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas G, Blanco E, Ballare C, Sanso M, Spill YG, Hu D, Aoi Y, Le Dily F, Shilatifard A, Marti-Renom MA, and Di Croce L (2018). Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat. Genet 50, 1452–1462. 10.1038/s41588-018-0218-5. [DOI] [PubMed] [Google Scholar]
- Modic M, Grosch M, Rot G, Schirge S, Lepko T, Yamazaki T, Lee FCY, Rusha E, Shaposhnikov D, Palo M, et al. (2019). Cross-regulation between TDP-43 and paraspeckles promotes pluripotency-differentiation transition. Mol. Cell 74, 951–965.e13. 10.1016/j.molcel.2019.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani S, Nichols J, and Hadjantonakis AK (2017). The many faces of Pluripotency: in vitro adaptations of a continuum of in vivo states. BMC Dev. Biol 17, 7. 10.1186/s12861-017-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet B, Roberts JM, Buckley DL, Paulk J, Dastjerdi S, Yang A, Leggett AL, Erb MA, Lawlor MA, Souza A, et al. (2018). The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol 14, 431–441. 10.1038/s41589-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, and Hirose T (2011). Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol 193, 31–39. 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F, Incarnato D, Krepelova A, Rapelli S, Pagnani A, Zecchina R, Parlato C, and Oliviero S (2013). Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol. 14, R91. 10.1186/gb-2013-14-8-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, and Smith A (2009). Naive and primed pluripotent states. Cell Stem Cell 4, 487–492. 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, and Chang HY (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Bi X, Pan Y, Gao B, Wu J, Yin Y, Liu Z, Peng M, Zhang W, Jiang X, et al. (2022). Phase separation of RNA-binding protein promotes polymerase binding and transcription. Nat. Chem. Biol 18, 70–80. 10.1038/s41589-021-00904-5. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, and Orkin SH (2008). EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502. 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalska L, Begley V, Beltran M, Lukauskas S, Khandelwal G, Faull P, Bhamra A, Tavares M, Wellman R, Tvardovskiy A, et al. (2021). Nascent RNA antagonizes the interaction of a set of regulatory proteins with chromatin. Mol. Cell 81, 2944–2959.e10. 10.1016/j.molcel.2021.05.026. [DOI] [PubMed] [Google Scholar]
- Smith A (2017). Formative pluripotency: the executive phase in a developmental continuum. Development 144, 365–373. 10.1242/dev.142679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. (2016). Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514. doi. 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, and Pasini D (2013). Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell 49, 645–656. 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- Verma N, Pan H, Dore LC, Shukla A, Li QV, Pelham-Webb B, Teijeiro V, Gonzalez F, Krivtsov A, Chang CJ, et al. (2018). TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nat. Genet 50, 83–95. 10.1038/s41588-017-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee WW, and Reinberg D (2013). A double take on bivalent promoters. Genes Develop. 27, 1318–1338. 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zuo H, Jin J, Lv W, Xu Z, Fan Y, Zhang J, and Zuo B (2019). Long noncoding RNA Neat1 modulates myogenesis by recruiting Ezh2. Cell Death Dis. 10, 505. 10.1038/s41419-019-1742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Goodrich KJ, Gooding AR, Naeem H, Archer S, Paucek RD, Youmans DT, Cech TR, and Davidovich C (2017a). Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell 65, 1056–1067.e5. 10.1016/j.molcel.2017.02.003. [DOI] [PubMed] [Google Scholar]
- Wang X, Paucek RD, Gooding AR, Brown ZZ, Ge EJ, Muir TW, and Cech TR (2017b). Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol 24, 1028–1038. 10.1038/nsmb.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xiang Y, Yu Y, Wang R, Zhang Y, Xu Q, Sun H, Zhao ZA, Jiang X, Wang X, et al. (2021). Formative pluripotent stem cells show features of epiblast cells poised for gastrulation. Cell Res. 31, 526–541. doi. 10.1038/s41422-021-00477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PAC, Rappsilber J, and Helin K (2011). TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348. 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Eve Sun Y, and Zhang Y (2011). Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393. 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang Y, Xu Q, Zhou C, Liu B, Du Z, Zhang K, Zhang B, Wang X, Gayen S, et al. (2020). Epigenomic analysis of gastrulation identifies a unique chromatin state for primed pluripotency. Nat. Genet 52, 95–105. 10.1038/s41588-019-0545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Zhang Z, Chen J, Huang H, Xu Y, Ding X, Zheng Y, Nishinakamura R, Xu GL, Wang H, et al. (2016). Cooperative action between SALL4A and TET proteins in stepwise oxidation of 5-methylcytosine. Mol. Cell 64, 913–925. 10.1016/j.molcel.2016.10.013. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, and Hirose T (2018). Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053.e7. 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- Yan J, Dutta B, Hee YT, and Chng WJ (2019). Towards understanding of PRC2 binding to RNA. RNA Biol. 16, 176–184. 10.1080/15476286.2019.1565283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z, Zhao Y, Shen B, Huang X, Zhu H, et al. (2015). Opposing roles forthe lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16,504–516. 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Yu L, Wei Y, Sun HX, Mahdi AK, Pinzon Arteaga CA, Sakurai M, Schmitz DA, Zheng C, Ballard ED, Li J, et al. (2021). Derivation of intermediate pluripotent stem cells amenableto primordial germ cell specification. Cell Stem Cell 28, 550–567.e12. 10.1016/j.stem.2020.11.003. [DOI] [PubMed] [Google Scholar]
- Zhu F, Zhu Q, Ye D, Zhang Q, Yang Y, Guo X, Liu Z, Jiapaer Z, Wan X, Wang G, et al. (2018). Sin3a-Tet1 interaction activates gene transcription and is required for embryonic stem cell pluripotency. Nucleic Acids Res. 46, 6026–6040. 10.1093/nar/gky347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq, ChIRP-seq, and RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) with accession code: GSE182443. The TET1 affinity purification followed by mass spectrometry data have been deposited at the ProteomeXchange Consortium via the PRIDE partner repository with accession code: PXD033587. The deposited data are publicly available as of the date of publication. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| TET1 | Millipore | Cat. 09-872; RRID: AB_10806199 |
| TET1 | GeneTex | Cat. GTX125888; RRID: AB_11164485 |
| PSPC1 | Santa Cruz | Cat. sc-84577; RRID: AB_2171459 |
| PSPC1 | Bethyl | Cat. A303-206A; RRID: AB_10954256 |
| PSPC1 | Sigma | Cat. SAB4200503; RRID: N/A |
| EZH2 | Cell Signaling | Cat. 5246S; RRID: AB_10694683 |
| SUZ12 | Abcam | Cat. ab12073; RRID: AB_442939 |
| SUZ12 | Active Motif | Cat. 39357; RRID: AB_2614929 |
| V5 | Invitrogen | Cat. R960-25; RRID: AB_2556564 |
| Mouse IgG | Millipore | Cat. 12-371; RRID: AB_145840 |
| Rabbit IgG | Millipore | Cat. PP64; RRID: AB_97852 |
| SIN3A | Abcam | Cat. ab3479; RRID: AB_303839 |
| PELP1 | Bethyl | Cat. A300-180A; RRID: AB_242526 |
| TET2 | Abcam | Cat. ab124297; RRID: AB_2722695 |
| NONO | Bethyl | Cat. A300-587A; RRID: AB_495510 |
| SFPQ | Abcam | Cat. ab38148; RRID: AB_945424 |
| HA | Abcam | Cat. ab9110; RRID: AB_307019 |
| OCT4 | Santa Cruz | Cat. sc-5279; RRID: AB_628051 |
| NANOG | Bethyl | Cat. A300-397A; RRID: AB_386108 |
| ESRRB | R&D Systems | Cat. PP-H6707; RRID: AB_2100411 |
| ACTIN | Sigma | Cat. A5441; RRID: AB_476744 |
| GAPDH | ProteinTech | Cat. 10494-1-AP; RRID: AB_2263076 |
| Histone3 | Abcam | Cat. ab1791; RRID: AB_302613 |
| H3K4me3 | EpiCypher | Cat. 13-0041; RRID: N/A |
| H3K27me3 | Cell Signaling | Cat. 9733S; RRID: AB_2616029 |
| VCL | Abcam | Cat. ab129002; RRID: AB_11144129 |
| Streptavidin-HRP | GE Healthcare | Cat. RPN1231 V; RRID: N/A |
| Mouse IgG HRP | Cell Signaling | Cat. 7076S; RRID: AB_330924 |
| Rabbit IgG HRP | Jackson ImmunoRes | Cat. 715-175-151; RRID: AB_2340820 |
| Trueblot Mouse IgG HRP | Rockland | Cat. 18-8817-31; RRID: AB_2610850 |
| Trueblot Rabbit IgG HRP | Rockland | Cat. 18-8816-31; RRID: AB_2610847 |
| DNA 5mC | Cell Signaling | Cat. 28692; RRID: AB_2798962 |
| DNA 5hmC | Active Motif | Cat. 39769; RRID: AB_10013602 |
| Anti-dsDNA | Abcam | Cat. ab27156; RRID: AB_470907 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | GIBCO | Cat. 11965-092 |
| Heat inactivated FBS | GIBCO | Cat. 35-011-CV |
| Penicillin-Streptomycin | GIBCO | Cat. 15140-122 |
| L-Glutamine | GIBCO | Cat. 25030-081 |
| MEM NEAA | GIBCO | Cat. 11140-050 |
| 2-Mercaptoethanol | Sigma | Cat. M6250 |
| Puromycin | Sigma | Cat. P9620-10ML |
| Hygromycin | Omega | Cat. HG-80 |
| N2 | GIBCO | Cat. 17502-048 |
| B27 | GIBCO | Cat. 17504-044 |
| DMEM/F-12 | GIBCO | Cat. 11-330-032 |
| Neurobasal | GIBCO | Cat. 21-103-049 |
| LIF | Lab prep | N/A |
| GSK3i (CHIR99021) | Sigma | Cat. SML1046-25MG |
| MEKi (PD0325901) | Selleckchem | Cat. S1036 |
| Recombinant Fgf2 | R&D System | Cat. 233-FB |
| Recombinant Activin A | R&D System | Cat. 338-AC |
| 13C615N4 L-arginine | Cambridge Isotope | Cat. CNLM-539-H |
| 13C615N2 L-lysine | Cambridge Isotope | Cat. CNLM-291-H |
| 13C6 L-lysine | Cambridge Isotope | Cat. CLM-2247-H |
| dTAG-13 | Tocris | Cat. 6605 |
| Deposited data | ||
| PSPC1 ChIP-seq in ESC | This paper | NCBI GEO: GSE182443 |
| SUZ12 ChIP-seq in ESC | This paper | NCBI GEO: GSE182443 |
| SUZ12 ChIP-seq in EpiLC upon Pspc1 KO, Neat1 KO, and Tet1-degron treatments | This paper | NCBI GEO: GSE182443 |
| H3K27me3 ChIP-seq in ESC and EpiLC upon Pspc1 KO, Neat1 KO, and Tet1-degron treatments | This paper | NCBI GEO: GSE182443 |
| H3K4me3 ChIP-seq in ESC and EpiLC upon Pspc1 KO and Neat1 KO | This paper | NCBI GEO: GSE182443 |
| Neat1 ChIRP-seq in WT and Neat1 KO ESC and EpiLC | This paper | NCBI GEO: GSE182443 |
| Pspc1 WT/KO RNA-seq in ESC and EpiLC | This paper | NCBI GEO: GSE182443 |
| Neat1 WT/KO RNA-seq in ESC and EpiLC | This paper | NCBI GEO: GSE182443 |
| TET1 affinity purification followed by mass spectrometry data in ESC | This paper | ProteomeXchange PRIDE: PXD033587 |
| TET1 ChIP-seq in ESC | Wu et al., 2011 | NCBI GEO: GSE26833 |
| PSPC1 CLIP-seq in ESC | Guallar et al., 2018 | NCBI GEO: GSE103269 |
| H3K4me3 and H3K27ac in ESC | Hon et al., 2014 | NCBI GEO: GSE48519 |
| 5mC and 5hmC meDIP-seq in ESC | Xiong et al., 2016 | NCBI GEO: GSE57700 |
| Experimental models: Cell lines | ||
| Mouse ESC CCE | This paper | N/A |
| Pspc1 KO ESC | Guallar et al., 2018 | N/A |
| Pspc1 KO ESC rescued with WT or RRMmut PSPC1 protein | Guallar et al., 2018 | N/A |
| Tet1-degron ESC | This paper | N/A |
| Mouse ESC 46C | This paper | N/A |
| Neat1 KO ESC | This paper | N/A |
| Ezh2 KO ESC | Shen et al., 2008 | N/A |
| Mouse ESC V6.5 | Laboratory of R. Jaenisch | N/A |
| Tet1 KO ESCs | Laboratory of R. Jaenisch | N/A |
| Tet1/2/3 KO ESCs | Laboratory of R. Jaenisch | N/A |
| Dnmt1/3a/3b KO ESC | Laboratory of T. Chen | N/A |
| Nono KO ESC | Laboratory of F. Lan | N/A |
| Oligonucleotides | ||
| Oligonucleotides (see Table S4) | This paper | N/A |
| Software and algorithms | ||
| STAR | 2.7.6a | https://github.com/alexdobin/STAR |
| Cufflinks | 2.2.1 | http://cole-trapnell-lab.github.io/cufflinks/ |
| Bowtie2 | 2.3.5 | http://bowtie-bio.sourceforge.net/bowtie2/ |
| IGV | 2.10.2 | https://software.broadinstitute.org/software/igv |
| samtools | 1.10 | http://www.htslib.org/ |
| PICARD | 2.18.5 | https://broadinstitute.github.io/picard/ |
| HOMER | 4.11.1 | http://homer.ucsd.edu/homer/ |
| MACS2 | 2.2.7 | https://github.com/macs3-project/MACS |
| NGSplot | 2.61 | https://github.com/shenlab-sinai/ngsplot |


