Figure 6. PSPC1, TET1, and Neat1 modulate PRC2 binding to nascent bivalent gene transcripts during bivalent gene activation.
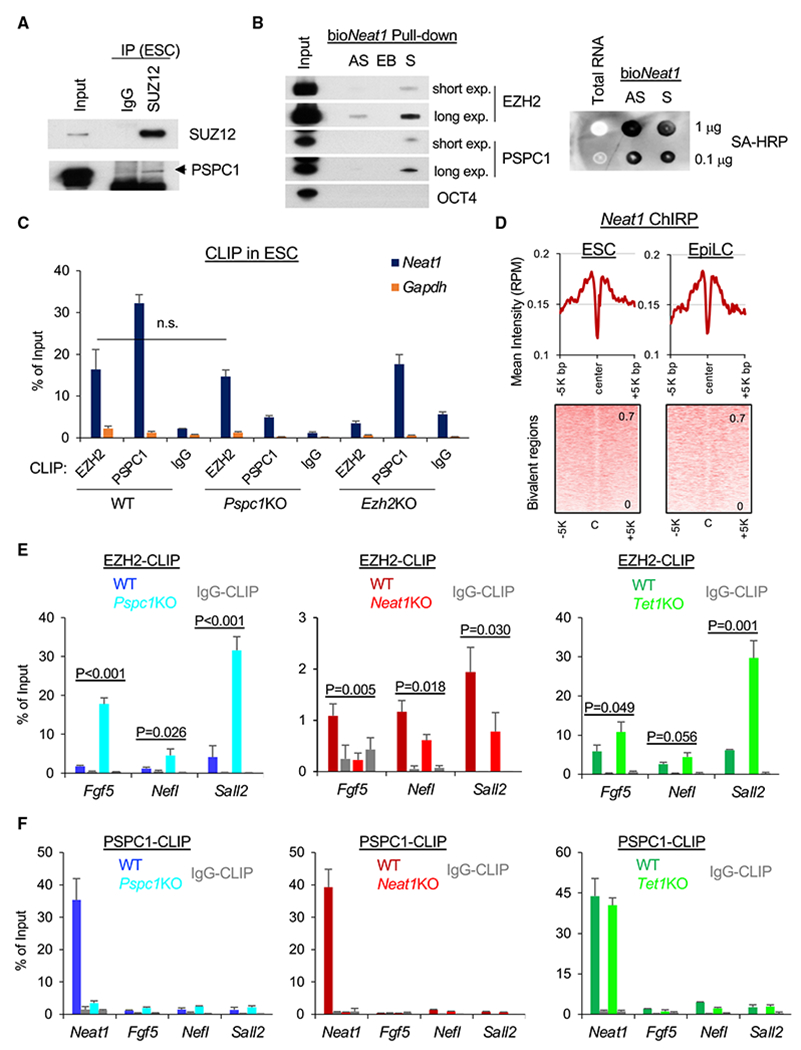
(A) Co-IP of PSPC1 and SUZ12 in ESCs using a nucleosome-containing protocol (see STAR Methods for detail).
(B) Biotinylated Neat1 (bioNeat1) RNAs pull down both EZH2 and PSPC1. Left: streptavidin (SA) beads conjugated with Neat1 sense (S) or antisense (AS) RNA, and empty beads(EB) were used for pull-down from ESC nuclear lysates followed by western blot analysis of bioNeat1-bound proteins. EZH2 and PSPC1 blots of both short and long exposure (exp.) are shown. Right: bioNeat1 sense (S) or antisense (AS) RNA were transcribed by in vitro transcription (IVT) and confirmed by SA-HRP dot blot. ESC total RNA serves as a negative control.
(C) EZH2 and PSPC1 CLIP-qPCR analysis of Neat1 in WT, Pspc1KO, and Ezh2KO ESCs. Gapdh serves as a negative control; p value is from two-tailed t test, and “n.s.” denotes statistically non-significant.
(D) Mean intensity plot (top) and heatmap (bottom) by RPM of Neat1 ChIP-seq intensity at the bivalent regions (within 5K bp at peak center, identified in ESCs) in ESCs and D2 EpiLCs.
(E) EZH2 CLIP-qPCR analysis of bivalent gene mRNAs (Fgf5, Nefl, and Sall2) in D2 EpiLCs of different genotypes (WT versus KO); p value is from the two-tailed t test.
(F) PSPC1 CLIP-qPCR analysis of Neat1 and bivalent genes’ transcripts (Fgf5, Neff, and Sall2) in D2 EpiLCs of different genotypes. Error bars in (C), (E), and (F) represent the standard deviation of technical triplicates. Experiments were repeated in biological duplicates.
