Figure 7. The working model of this study.
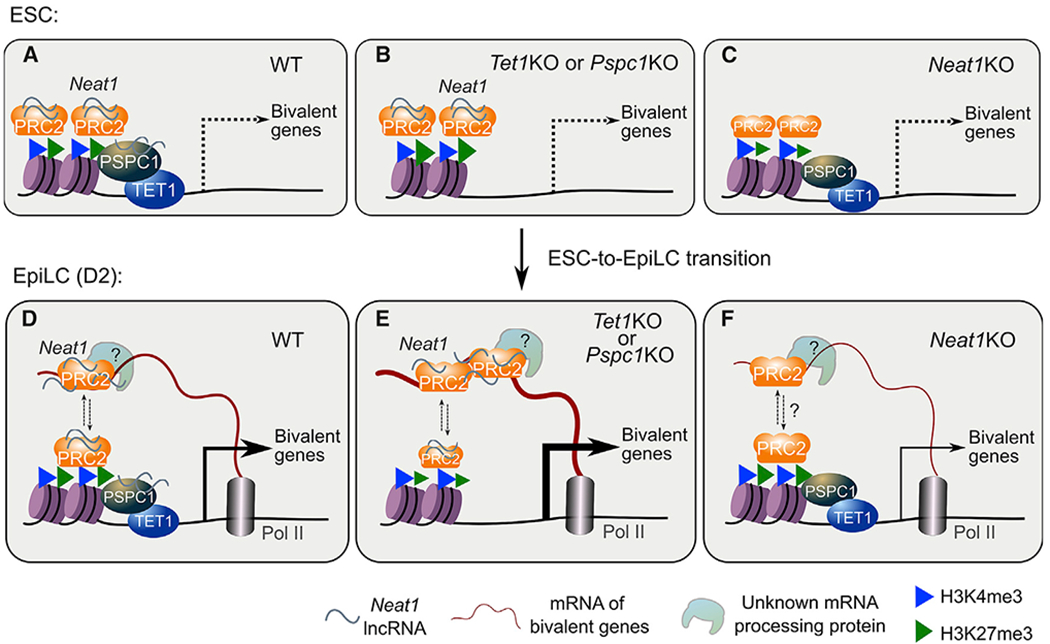
(A–C) In ESCs (WT), Neat1 (short isoform, Neat1_1) associates with the chromatin-bound proteins TET1, PSPC1, and PRC2 at bivalent gene promoters (A). Bivalent genes are minimally expressed in WT (A), Tet1KO, or Pspc1KO (B), or Neat1KO (C) ESCs. In Neat1KO ESCs and D2 EpiLCs(WT or KO), the chromatin-bound PSPC1 and TET1 decrease, denoted by smaller protein symbols. Bivalent genes are activated during pluripotent-state transition (accompanied by downregulation of Neat1_1, with no expression of Neat1_2 yet), and nascent mRNA acts as a decoy to evict PRC2 from chromatin.
(D–F) In EpiLCs(WT), a dynamic balance is maintained between PRC2 chromatin occupancy and RNA binding (shown in up/down arrows)to fine-tune the expression of bivalent genes (D). In Tet1KO or Pspc1KO (E) EpiLCs, more PRC2 proteins bind to mRNAs and are displaced or evicted from chromatin, inducing enhanced bivalent gene transcription. Without Neat1 (F), the balance between the chromatin- and mRNA-bound PRC2 may be disrupted (indicated by dashed lines and a question mark). PRC2-binding affinityto mRNAs (and possibly mRNA-processing-associated proteins) is compromised, which causes reduced bivalent gene activation. Of note, although PRC2 binds to both Neat1 and certain bivalent gene transcripts, Neat1 may promote PRC2 binding to nascent mRNA transcripts indirectly (D–F, e.g., through unknown mRNA-processing protein; see Limitations of the study).
