Dear Editor,
Autophagy is an evolutionarily conserved catabolic process that involves the sequestration and transport of organelles, macromolecules, or invading microorganisms to lysosomes for degradation [1]. Sequestosome 1 (p62/SQSTM1) was the first protein shown to bind target-associated ubiquitin (Ub) and LC3 conjugated to the phagophore membrane, thus, acting as an important autophagy receptor for ubiquitinated targets [2].
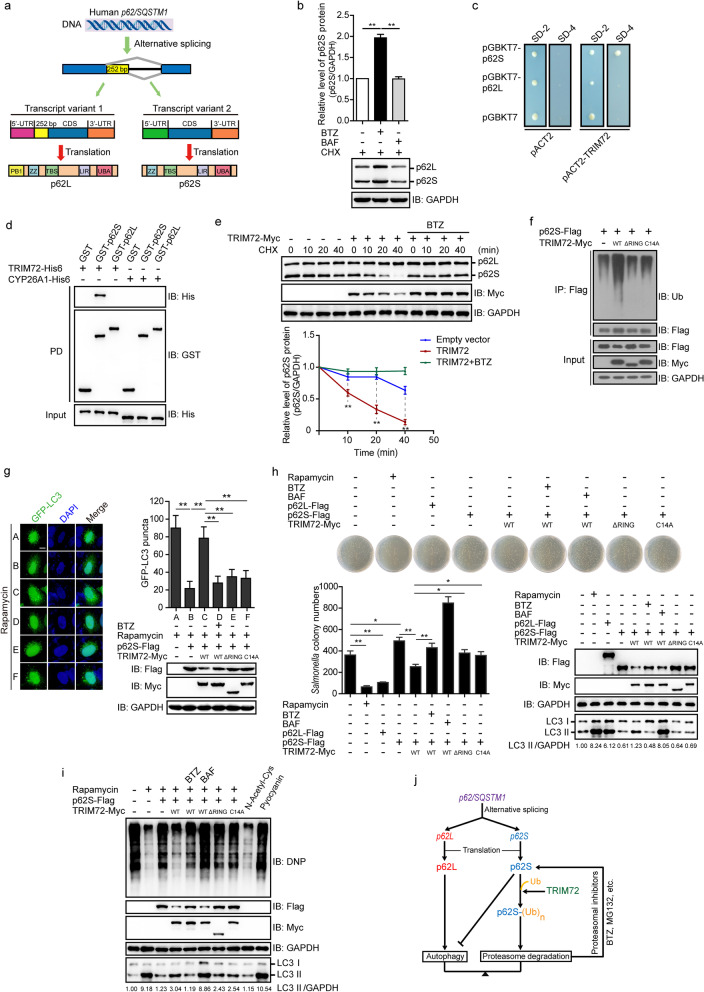
Human p62/SQSTM1 has alternative 5’donor sites during splicing of immature mRNA, which results in two isoforms of the p62/SQSTM1 transcripts, p62L and p62S. In contrast to p62L, differences in the 5’-untranslated region (UTR) and coding sequence (CDS) region between the two transcript variants resulted in a loss of 1–84 amino acids (included within the PB1 domain) in p62S (Fig. 1a, Additional file 1: Fig. S1a). In this study, two pairs of primers were designed to specifically amplify the cDNA fragments of p62 isoforms in different cell lines (Additional file 1: Table S1). Expression of p62L was detected in all 9 cell lines, whereas p62S expression was detected in most cell lines, with the exception of human skeletal muscle cells (HSkMC) and adult hepatocytes (Additional file 1: Fig. S1b). No p62S expression was found in various tissues of mice (Additional file 1: Fig. S1c), indicating a human cell-type-specific expression pattern in the biogenesis of p62S. An shRNA specifically targeting p62S but not p62L was designed (Additional file 1: Table S2), and validated by quantitative PCR and immunoblot analysis (Additional file 1: Fig. S1d), indicating the presence of p62S.
Fig. 1.
TRIM72-mediated ubiquitin signaling controls the homeostasis of the p62/SQSTM1 short isoform in regulating selective autophagy. a Schematic domain structure of p62L and p62S, which were derived from alternative splicing of the human p62/SQSTM1 gene. b Endogenous p62S and p62L were mainly degraded by proteasomes, but not by the autophagy pathway in HeLa cells treated with bortezomib (BTZ, 1 μmol/L) or bafilomycin (BAF, 20 nmol/L), as well as cycloheximide (CHX, 100 μg/ml). c Yeast two-hybrid screening identified TRIM72 as an interacting partner for p62S, but not for p62L. SD-2 was deficient in Leu and Trp, and SD-4 was deficient in Ura, His, Leu, and Trp. d Recombinant glutathione-S-transferase (GST)-tagged p62S directly interacted with His6-tagged TRIM72 in vitro as detected by a GST pull-down (PD) assay. Recombinant His6-tagged CYP26A1 acted as a negative control. e TRIM72 promoted the degradation of endogenous p62S, but not p62L, using the proteasome pathway. HeLa cells were ectopically expressed with empty vector or TRIM72, treated with the indicated compounds during different times, and detected by immunoblot analyses. f Wild-type TRIM72 but not the E3 ligase death mutants (TRIM72ΔRING and TRIM72C14A) supported the poly-ubiquitylation of p62S. HEK293T cells were ectopically expressed with the indicated plasmids, and then the cell lysates were immunoprecipitated with anti-Flag affinity gels before being subjected to immunoblotting analysis. g HeLa cells were co-transfected with the indicated plasmids, treated with 2 μmol/L rapamycin for 12 h, then with or without BTZ for 1 h, and subjected to fluorescent microscopy. Puncta formation by GFP-LC3 were counted and calculated. The protein levels of TRIM72 and p62S were also detected by immunoblot analysis. Scale bar = 10 μm. h Salmonella infection assay indicated that TRIM72 mediated the proteasomal degradation of p62S, and facilitated the clearing of Salmonella. HeLa cells were transfected with the indicated plasmids for 24 h, and then treated with the indicated compounds before Salmonella infection for 30 min. The cells were then lysed and one-tenth of the lysate was subjected to plate assays. The Salmonella colony numbers were counted and calculated. The protein levels of TRIM72, p62L, p62S, and LC3 were also detected by immunoblot analysis, and the expression of lipidated LC3 (LC3 II) was quantitated after normalization of control values as 1.00. i TRIM72-mediated proteasomal degradation of p62S facilitated carbonylated protein degradation. HeLa cells were transfected with the indicated plasmids for 24 h, and then treated with the indicated compounds before being subjected to the detection of protein oxidation. Pyocyanin was used as a positive control, and N-acetyl-l-cysteine was used as a negative control. Carbonylated proteins were visualized after derivatization with 2,4-dinitrophenylhydrazine (DNPH), followed by immunoblotting with anti-DNP. The levels of lipidated LC3 (LC3 II) were quantitated after normalization of the control as 1.00. j A model showing how TRIM72-mediated proteasomal degradation of p62S rheostatically regulated selective cellular autophagy. Data are presented as the mean ± SD, and analyzed with one-way analysis of variance using the Bonferroni post-hoc test or the two-tailed unpaired t test. *P < 0.05, **P < 0.01, three independent experiments. CDS coding sequence, CHX cycloheximide, Myc an epitope tag for protein tagged, TRIM72 tripartite motif-containing 72, Ub ubiquitin, UTR Untranslated region
p62S and p62L protein levels were upregulated when treated with the proteasome inhibitor bortezomib (BTZ), but not with the autophagy inhibitor bafilomycin (BAF, Fig. 1b; Additional file 1: Fig. S2a, b), indicating that p62S and p62L were mainly degraded by the proteasome pathway rather than the autophagy pathway.
Using the yeast two-hybrid screening system, tripartite motif-containing 72 (TRIM72) was identified as interacting with p62S but not with p62L (Fig. 1c). The interaction of TRIM72 and p62S was subsequently verified by glutathione-S-transferase (GST)-pull-down (Fig. 1d) and co-immunoprecipitation assays (Additional file 1: Fig. S3a). TRIM72 promoted the degradation of p62S in a dose-dependent manner (Additional file 1: Fig. S3b), and ectopic expression of TRIM72 decreased p62S protein level and increased lipidated LC3 (LC3 II) level, while TRIM72 knockdown increased p62S level and decreased LC3 II level (Additional file 1: Fig. S3c). Pulse-chase experiments revealed that p62S, but not p62L protein, was reduced upon TRIM72 overexpression, indicating a specific effect of TRIM72 on p62S proteasomal degradation (Fig. 1e). Furthermore, wild-type TRIM72, but not an E3 ligase activity death mutant (TRIM72ΔRING or TRIM72C14A), supported the ubiquitination of p62S (Fig. 1f). Together, these results suggest that TRIM72 is a p62S-specific E3 ligase.
Regarding the physiological function of the TRIM72-mediated degradation of p62S protein, PB1 lacking p62S bound to the poly-ubiquitinated cargo and was capable of binding to LC3 as efficiently as p62L (Additional file 1: Fig. S4a, b). Upon treatment with rapamycin, p62S reduced GFP-LC3 puncta formation in HeLa cells, indicating that p62S suppressed rapamycin-induced autophagy, which was reversed by wild-type TRIM72 but not its mutants (TRIM72ΔRING and TRIM72C14A, Fig. 1g). In addition, TRIM72 regulated autophagy, mainly through p62S, but not through p62L, as shown in p62−/− MEF cells (Additional file 1: Fig. S4c).
Because invading microorganisms are mainly degraded through autophagy, a Salmonella infection assay was used to measure cellular autophagic activities [2]. Introduction of rapamycin and p62L into HeLa cells reduced the number of Salmonella infection by increasing autophagy (LC3 II), while introduction of p62S increased the number of infected Salmonella by reducing autophagy. When p62S was down-regulated by wild-type TRIM72, the number of infected Salmonella were significantly reduced due to increased autophagy (Fig. 1h).
Another function of cellular autophagy is to remove proteins from oxidative damaged (carbonylation) [3]. The degradation of carbonylated proteins in HeLa cells after rapamycin treatment was accelerated by increasing autophagy (LC3 II). This process was reduced in the presence of p62S, and the introduction of wild-type TRIM72 almost abolished the inhibitory effect of p62S, which was abolished by BTZ (Fig. 1i).
Taken together, our results revealed a previously unexplored mechanism involving alternative splicing of p62/SQSTM1 mRNA with TRIM72-mediated Ub signaling, to rheostatically control the cellular autophagic flux (Fig. 1j). Further exploitation of this mechanism offers new opportunities to develop therapeutic interventions for these related pathophysiological processes.
Supplementary Information
Additional file 1: Materials and Methods. Fig. S1 Human p62 has a shorter isoform p62S lacking the PB1 domain at the N-terminus. Fig. S2 Human p62S is mainly degraded though the proteasome pathway. Fig. S3 Human E3 ligase TRIM72 mediates the ubiquitination and degradation of p62S-regulated cellular autophagy. Fig. S4 Human p62S antagonizes the autophagy receptor function of p62L. Table S1 Sequences of the primers used in RT-PCR and qPCR. Table S2 Sequences of the shRNAs for p62S and TRIM72.
Acknowledgements
We thank all members of our laboratory for their assistance.
Abbreviations
- BTZ
Bortezomib
- BAF
Bafilomycin
- CDS
Coding sequence
- GST
Glutathione-S-transferase
- HSkMC
Human skeletal muscle cell
- LC3
Microtubule-associated protein 1A/1B-light chain 3
- SQSTM1/p62
Sequestosome 1
- TRIM72
Tripartite motif-containing 72
- Ub
Ubiquitin
- UTR
Untranslated region
Author contributions
RGH, CYL, and WJG conceived, designed, and supervised the project. CCW, HP, CYL, and JY performed most of the experiments. RGH, CYL, and ZW wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the Ministry of Science and Technology of China (2019YFA0802103), the National Natural Science Foundation of China (31900804, 31960179), the Department of Science and Technology of Zhejiang Province (2021C03104), and the Science and Technology Commission of Shanghai Municipality (19140903500).
Availability of data and materials
All the data and materials generated or analyzed in this study are available for other researchers after the manuscript is published.
Declarations
Ethics approval and consent to participate
Animal experiments were performed in compliance with guidance for the care and use of laboratory animals, and were approved by the Institutional Animal Research Ethics Committee of Shanghai Institute of Biochemistry and Cell Biology (2019-012).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Cheng-Cheng Wang and Hong Peng contributed equally to this work.
Contributor Information
Cheng-Cheng Wang, Email: wang.chengcheng22@163.com.
Hong Peng, Email: pengh37@mail.sysu.edu.cn.
Zi Wang, Email: wangzi0421@126.com.
Jiao Yang, Email: yangjiao@sibcb.ac.cn.
Rong-Gui Hu, Email: coryhu@sibcb.ac.cn.
Chuan-Yin Li, Email: lichuanyin2013@sibcb.ac.cn.
Wu-Jun Geng, Email: gengwujun@126.com.
References
- 1.Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) Autophagy. 2021;17(1):1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng H, Yang J, Li G, You Q, Han W, Li T, et al. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res. 2017;27(5):657–674. doi: 10.1038/cr.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Z, Chen P, Gao H, Gu Y, Yang J, Peng H, et al. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell. 2014;26(1):106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Materials and Methods. Fig. S1 Human p62 has a shorter isoform p62S lacking the PB1 domain at the N-terminus. Fig. S2 Human p62S is mainly degraded though the proteasome pathway. Fig. S3 Human E3 ligase TRIM72 mediates the ubiquitination and degradation of p62S-regulated cellular autophagy. Fig. S4 Human p62S antagonizes the autophagy receptor function of p62L. Table S1 Sequences of the primers used in RT-PCR and qPCR. Table S2 Sequences of the shRNAs for p62S and TRIM72.
Data Availability Statement
All the data and materials generated or analyzed in this study are available for other researchers after the manuscript is published.