ABSTRACT
Hepatitis C virus (HCV) is characterized by a high number of chronic cases owing to an impairment of innate and adaptive immune responses. CD81 on the cell surface facilitates HCV entry by interacting with the E2 envelope glycoprotein. In addition, CD81/E2 binding on immunity-related cells may also influence host response outcome to HCV infection. Here, we performed site-specific amino acid substitution in the front layer of E2 sequence to reduce CD81 binding and evaluate the potential of the resulting immunogen as an HCV vaccine candidate. The modified sE2 protein (F442NYT), unlike unmodified sE2, exhibited a significant reduction in CD81 binding, induced higher levels of proinflammatory cytokines, repressed anti-inflammatory response in primary monocyte-derived macrophages as antigen-presenting cells, and stimulated CD4+ T cell proliferation. Immunization of BALB/c mice with an E1/sE2F442NYT nucleoside-modified mRNA-lipid nanoparticle (mRNA-LNP) vaccine resulted in improved IgG1-to-IgG2a isotype switching, an increase in neutralizing antibodies against HCV pseudotype virus, a B and T cell proliferative response to antigens, and improved protection against infection with a surrogate recombinant vaccinia virus-expressing HCV E1-E2-NS2aa134-966 challenge model compared to E1/unmodified sE2 mRNA-LNP vaccine. Further investigation of the modified E2 antigen may provide helpful information for HCV vaccine development.
IMPORTANCE Hepatitis C virus (HCV) E2-CD81 binding dampens protective immune response. We have identified that an alteration of amino acids in the front layer of soluble E2 (sE2) disrupts CD81 interaction and alters the cytokine response. Immunization with modified sE2F442NYT (includes an added potential N-linked glycosylation site and reduces CD81 binding activity)-mRNA-LNP candidate vaccine generates improved proinflammatory response and protective efficacy against a surrogate HCV vaccinia challenge model in mice. The results clearly suggested that HCV E2 exhibits immunoregulatory activity that inhibits induction of robust protective immune responses. Selection of engineered E2 antigen in an mRNA-LNP platform amenable to nucleic acid sequence alterations may open a novel approach for multigenotype HCV vaccine development.
KEYWORDS: hepatitis C virus, modified E2, proinflammatory response, vaccine antigen selection, vaccine antigen
INTRODUCTION
HCV causes chronic liver disease in many infected humans and is one of the contributing factors for cirrhosis and hepatocellular carcinoma. Direct-acting antiviral agents (DAAs) have transformed the standard therapy, achieving high rates of sustained virologic response. Unfortunately, despite antiviral efficacy, DAAs cannot fully eradicate liver cancer risk, especially after HCV clearance in the context of advanced liver disease (1).
HCV has evolved mechanisms to evade immune activation and resolution of infection. Several studies of acute HCV infection demonstrate that virus clearance is associated with broad and potent T cell activation (2), and a rapid induction of cross-reactive neutralizing antibody responses (3). In a phase I vaccine trial of recombinant HCV E1/E2 envelope glycoproteins in healthy human volunteers, participant peripheral blood mononuclear cells (PBMCs) exhibited increased secretion of interleukin-10 (IL-10), an anti-inflammatory cytokine, and generated a variable range of IL-4 secretion when immunized with the candidate HCV E1/E2 vaccine. Vaccination failed to exhibit clear dose-dependent protective immune parameters (proinflammatory cytokine production, lymphoproliferative response, and neutralizing antibody response) to escalated E1/E2 protein administration, and only ~28% of the vaccinated human sera had a detectable virus neutralization response (4). Thus, HCV E1/E2 did not appear to be very effective as a candidate vaccine for induction of a robust protective immune response. A recent report on a clinical vaccine trial administering a recombinant viral vectored vaccine expressing nonstructural proteins showed vaccination resulted in lower HCV RNA levels but did not prevent chronic infection (5).
We have subsequently shown that HCV E2 induces the immune regulatory cytokines IL-10 and CD163 protein from primary macrophages (6). Further, HCV E2 enhances STAT3 and suppresses STAT1 activation, suggesting macrophage polarization toward the M2 phenotype. HCV E2 interrupts the function of T, B, and NK cells by binding with CD81 (7–9). HCV-specific CD4+ T cell immune deficiency may be the primary cause of CD8+ T cell functional exhaustion (10).
HCV E2 contains multiple epitopes for both T and B cells (11). Here, we examine whether impairing the E2-CD81 interaction can promote T-helper cell functions to induce a robust HCV E2 antigen- specific immune response. Further, to examine the nature of immunogenicity of the modified E2 antigens in comparison to wild-type sE2, we took advantage of utilizing a mRNA-LNP vaccine platform in a preclinical mouse model and a recombinant vaccinia virus expressing HCV proteins for challenge as a surrogate model for clearance. Our results suggest that reduced HCV E2-CD81 interaction significantly improves immune responses. These findings may contribute significantly to the development of an effective HCV vaccination strategy.
(This manuscript was deposited as a preprint in bioRxiv [12].)
RESULTS
Amino acid substitution in E2 front layer impairs CD81 binding.
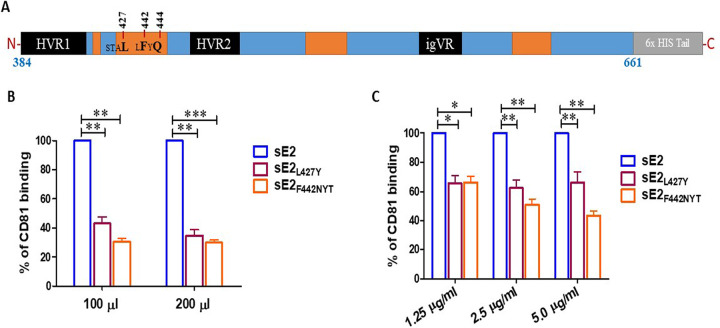
The front layer of the HCV E2 glycoprotein interacts with the large extracellular loop of CD81 in the process of viral entry (13). The front layer is located within two hypervariable regions of the HCV E2 ectodomain and exhibits flexible antigenic activity (14). We have postulated two different approaches from an earlier report (15) on structural and CD81 binding analyses of E2 glycoprotein to design variants to disrupt CD81 interaction in HCV genotype 1a H77 strain lacking the hydrophobic C-terminal transmembrane (amino acids 384 to 661) domain (Fig. 1A). For one such approach, we selected nucleotide change for one amino acid substitution by replacing nonpolar leucine with polar tyrosine at position 427 (L427Y). In another approach, we introduced nucleotide changes for a N-linked glycosylation site (N-X-T/S) at positions 442 on the front layer of E2 (F442NYT) to abrogate E2-CD81 binding containing an additional N-linked glycosylated site. We investigated the effect of modified E2 on CD81 in a qualitative binding assay. sE2 or modified E2 were expressed in mammalian HEK293T cells. We tested the E2-CD81 interaction of different E2 variant proteins from transfected cell culture medium by enzyme-linked immunosorbent assay (ELISA). A significant reduction in CD81 binding for cell culture derived sE2L427Y (~60% loss) and sE2F442NYT (~70% loss) compared to the unmodified sE2 protein was observed under our experimental conditions (Fig. 1B). Secreted sE2 and modified E2 bearing a C-terminal hexahistidine tag were purified from culture supernatants by immobilized nickel affinity chromatography. Purified protein from sE2L427Y and sE2F442NYT also exhibited decreases in binding activity to ~35% and ~40%, respectively at 2.5 μg/ml E2 concentration (Fig. 1C). Thus, amino acid substitutions in the front layer of CD81 binding domain of E2 resulted in an impairment of CD81 interaction.
FIG 1.
Schematic presentation of HCV sE2 ectodomain and the mutants. (A) Amino acid positions of interest for mutations in a linear map of sE2 are indicated. Orange boxes indicate CD81 binding determinants. (B and C) Comparison of human CD81 binding with modified E2 expressed transiently in HEK293T-transfected cell culture supernatant with increasing volumes and purified histidine tag recombinant proteins in a dose-dependent concentration. The culture medium from mock-transfected HEK293T cells or phosphate-buffered saline was used as a reagent control. The significance level is indicated (*, P < 0.05; **, P < 0.005; ***, P < 0.001).
Inhibition of E2/CD81 binding induces proinflammatory cytokine production.
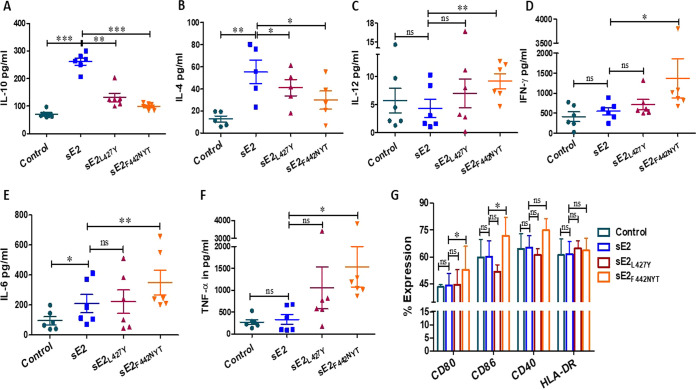
Both macrophages and dendritic cells (DCs) play an important role in immune surveillance. We have previously shown that HCV E2 significantly inhibits macrophage polarization toward the M1 phenotype, and the maturation of antigen-presenting DCs (6, 16, 17). HCV E2 binds to its cognate receptor, CD81, on macrophages and induces the production of IL-10. We analyzed the immune modulatory effect of modified E2 on primary human monocyte-derived macrophages. sE2 induced significantly higher IL-10 generation in macrophages compared to the modified sE2 (sE2L427Y or sE2F442NYT) proteins (Fig. 2A). Within the modified sE2 group, the sE2L427Y generated more IL-10 compared to sE2F442NYT. Similar to what was seen with IL-10, a significant increase in the production of IL-4, which induces differentiation of naive T cells to the Th2 phenotype, was observed in macrophages incubated with the sE2 protein, and this effect was mitigated with the modified sE2 proteins (Fig. 2B). IL-10 may impair the induction of gamma interferon (IFN-γ) and the associated cytokines IL-12 and tumor necrosis factor alpha (TNF-α) (18). We observed a significant increase in proinflammatory cytokine production with modified sE2 compared to unmodified sE2 and unstimulated control macrophages. Among the tested proinflammatory cytokines, IL-12 induction was consistently higher in macrophages exposed to modified sE2F442NYT proteins (Fig. 2C), while exposure of macrophages to unmodified sE2 generated little production of IFN-γ above that of the control (Fig. 2D). We measured a significant increase in IFN-γ levels from macrophages exposed to the sE2F442NYT protein. Within the treatment groups, sE2F442NYT produced significantly higher IL-6 levels compared to sE2L427Y and sE2 (Fig. 2E). We also observed a significantly greater TNF-α generation from sE2F442NYT protein (Fig. 2F). These results suggest that as the E2/CD81 interaction diminishes, the ratio of proinflammatory to anti-inflammatory cytokine expression increases.
FIG 2.
Expression of cytokine and costimulatory molecules from macrophages of healthy donors incubated with sE2 or modified E2 for comparison. (A to F) Monocyte-derived macrophages from healthy donors (n = 6) were incubated with 2.5 μg/mL HCV sE2 or modified proteins for 24 h. Secretory cytokines (IL-10, IL-4, IL-12, IFN-γ, IL-6, and TNF-α) in the culture supernatant were quantified by ELISA. (G) The expression of costimulatory molecules on the surfaces of activated macrophages was quantified by flow cytometry. All results were compared to blank controls. The significance level is indicated (*, P < 0.05; **, P < 0.005; ***, P < 0.001; ns, not significant).
M1 macrophages are characterized by high IL-12 and low IL-10 expression and express high levels of T cell-related costimulatory molecules CD80, CD86, and CD40 for the activation of T cells (19, 20). We tested the ability of sE2, sE2L427Y, and sE2F442NYT proteins to induce expression of the activation markers CD80, CD86, CD40, and HLA-DR on monocyte-derived macrophages from healthy donors in the presence or absence of recombinant proteins incubated for 24 h. The expression of the activation markers was measured by flow cytometry. We found significantly increased CD80 and CD86 expression in sE2F442NYT-stimulated macrophages compared to unstimulated, sE2, or sE2L427Y macrophages (Fig. 2G). We observed higher, although not statistically significant, CD40 expression and unaltered HLA-DR expression upon sE2F442NYT treatment. Taken together, the results indicate that the presence of sE2F442NYT induces a profile in macrophages associated with the classical M1 phenotype.
sE2F442NYT-activated macrophages promote CD4+ T cell differentiation into Th1 type.
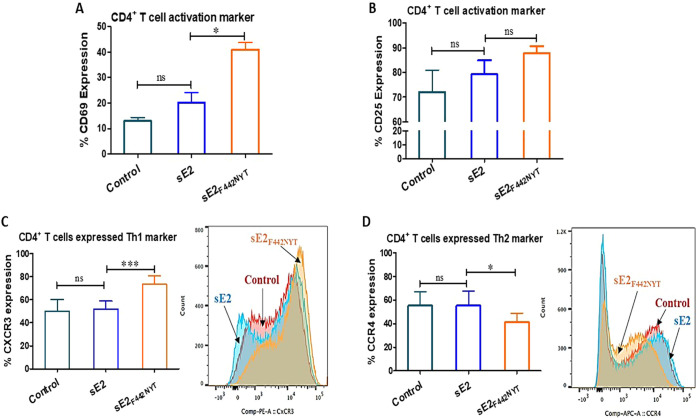
Differentiation of naive T cells into the Th1 effector type is critical to induce a protective immune response against intracellular pathogens (21). Polarization of Th0 into Th1- or Th2-type effectors depends on the type and concentration of cytokines present at the immune synapse, the strength of the T cell receptor-mediated signals, and the activation state of the antigen-presenting cells. Based on our results, we hypothesized that the activated macrophages could polarize T cells toward Th1-type effector cells. To test our hypothesis, we incubated macrophages in the presence or absence of sE2 or sE2F442NYT for 24 h and cocultured them with autologous CD4+ T cells. CD69 surface expression correlates with differentiation of CD4+ T cells into Th1 effector cells (22). We observed significant increased expression of the CD69 activation marker in both sE2F442NYT-treated cells compared to sE2-treated CD4+ T cells (Fig. 3A). CD25 is the α-chain of the IL-2 receptor and a T cell activation marker that responds to proinflammatory cytokines. We observed higher CD25 surface expression in cells cocultured with sE2- or sE2F442NYT-treated macrophages for 24 h (Fig. 3B). A significant increase was noted in the Th1 marker, CXCR3, on CD4+ T cells cocultured with sE2F442NYT compared to untreated or sE2-treated macrophages (Fig. 3C). We found a significant decrease in the expression of the Th2 marker CCR4 on CD4+ T cells upon sE2F442NYT treatment (Fig. 3D). These results support the finding that macrophages stimulated with sE2F442NYT promote differentiation of CD4+ T cells toward a Th1 type compared to unmodified sE2.
FIG 3.
Effect of macrophage activation by sE2F442NYT on the polarization of CD4+ T cells toward a Th1 type. (A and B) CD4+ T cells were isolated from healthy donors (n = 4) and treated with sE2 or sE2F442NYT for 2 h in the presence of human anti-CD3 (5 μg/mL) and anti-CD28 (1 μg/mL). The expression of T cell activation markers CD69 and CD25 on the surfaces of CD4+ T cells was quantified by flow cytometry. (C and D) Monocyte-derived macrophages isolated from healthy donors (n = 6) were treated with sE2 or sE2F442NYT protein for 24 h, and autologous CD4+ T cells were cocultured with the treated macrophages for 4 days. Th1 and Th2 polarization was measured as the expression of CXCR3 and CCR4, respectively, on the surfaces of CD4+ T cells by flow cytometry. The significance level is indicated (*, P < 0.05; ***, P < 0.001; ns, not significant).
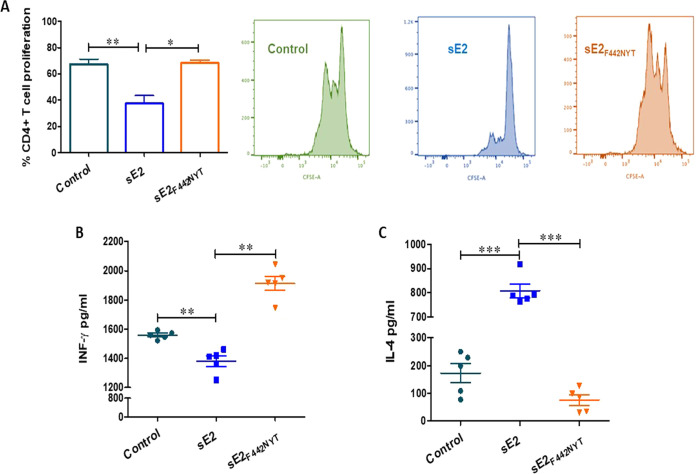
Proliferative CD4+ T cell response against HCV proteins is a strong immunological correlate of the outcome of acute HCV infection (23) and are usually not detectable in acute persisting and chronic infection. Here, we analyzed the role of modified E2 in promoting isolated CD4+ T cell proliferation. CD4+ T cells isolated from healthy volunteers were carboxyfluorescein succinimidyl ester (CFSE) stained and incubated with purified sE2 or sE2F442NYT for 72 h. Fluorescence-activated cell sorting (FACS) analyses revealed a significant impairment in CD4+ T cell proliferation following sE2 treatment (Fig. 4A). In contrast, sE2F442NYT-treated cells displayed a similar proliferation response as a control.
FIG 4.
Evaluation of human CD4+ T cell response isolated from healthy donors stimulated with HCV envelope E2 antigens. (A) CD4+ T cells isolated by magnetic bead separation were incubated with purified sE2 or sE2F442NYT for 72 h in the presence of CFSE, and the proliferation results from FACS analyses are shown. (B) IFN-γ and (C) IL-4 were analyzed from culture supernatants of CD4+ T cells responding to stimulation with sE2 or sE2F442NYT after 72 h. The significance level is indicated (*, P < 0.05; **, P < 0.005; ***, P < 0.001; ns, not significant).
We evaluated the potential for T cell expansion from secretion of IFN-γ under the influence of HCV wild-type or modulated sE2 antigen for further T cell functional evaluation. On the other hand, IL-4 is known to promote the differentiation of CD4+ T cells into IL-4-secreting Th2 cells. IFN-γ and IL-4 were analyzed separately in CD4+ T cells responding to stimulation in the presence of sE2 or sE2F442NYT (Fig. 4B). sE2 decreased IFN-γ production in CD4+ T cells but showed an increase in IL-4 production. In contrast, under the same treatment conditions, sE2F442NYT did the opposite and increased IFN-γ production by 80% over the positive control and decreased IL-4 production. Thus, the results suggest that the sE2F442NYT variant reverses the response of CD4+ T cells exhibited by unmodified sE2.
Immunization of mice with E1/sE2F442NYT mRNA-LNP vaccine induces Th1-related cytokines.
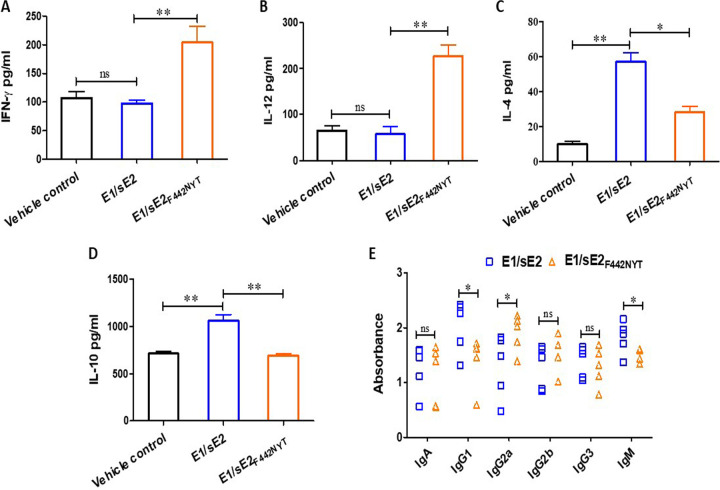
The lack of a suitable small animal model for use as a host for HCV infection hinders evaluation for vaccine evaluation in a natural host to challenge study. Here, we tested the immune stimulatory effect of sE2 variants in mice immunized with E1/sE2 or E1/sE2F442NYT and protection from surrogate HCV vaccinia challenge. We observed a significant increase in IFN-γ and IL-12 expression in E1/sE2F442NYT-vaccinated mice compared to E1/sE2 and vehicle control mice (Fig. 5A and B). Within treatment groups, E1/sE2F442NYT produced significantly less IL-4 compared to E1/sE2 (Fig. 5C). The E1/sE2 construct induced significantly higher IL-10 production in serum compared to the E1/sE2F442NYT construct, and this was highly significant compared to the vehicle control (Fig. 5D). This observation supports our earlier findings that the ablation of E2/CD81 interaction in macrophages reduces E2-induced IL-10 production. Antibody isotype switching from IgM to IgG isotypes occurs after antigen exposure. The Th1 subtype of CD4+ T cells secrete IFN-γ, which induces an isotype switch in B cells to IgG2a secretion, whereas Th2 cells are associated with enhanced IgG1 production. Here, mice immunized with the E1/sE2-mRNA vaccine preparation exhibited pronounced IgG1 in serum. In contrast, a distinct skew toward IgG2a was apparent in the isotypes of the E1/sE2F442NYT mRNA-immunized mice (Fig. 5E). Taken together, these data show that immunization of mice with the E1/sE2F442NYT mRNA vaccine promotes a Th1 response.
FIG 5.
Comparison of mouse cytokine response after immunization with E1/sE2 or E1/sE2F442NYT-mRNA-LNP candidate vaccine. (A to D) IFN-γ, IL-12, IL-4 and IL-10 levels were quantified by ELISA in the sera of vehicle control, E1/sE2-vaccinated, or E1/sE2F442NYT-vaccinated mice. (E) Immunized mouse sera (collected after 4 weeks) were analyzed for the isotype-specific antibody response by coating purified E2 antigen on commercially available ELISA plate for isotype antibody analyses. Results from individual sera are shown from different immunized groups of mice. The significance level is indicated (*, P < 0.05; **, P < 0.005; ***, P < 0.001; ns, not significant).
E1/sE2F442NYT mRNA-LNP vaccine enhances protective immune response.
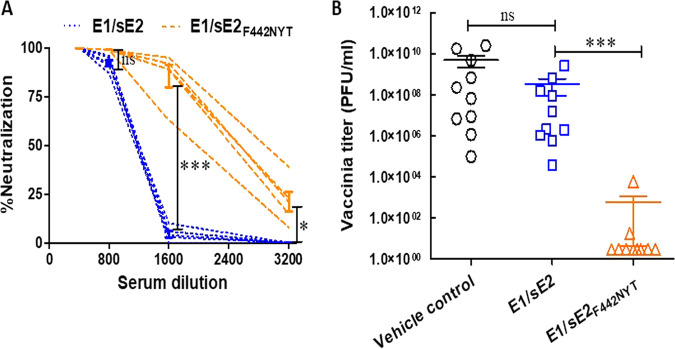
An HCV-lentiviral pseudotype particle (HCVpp) system was used to analyze relative neutralization activity between these two immunogen preparations. Sers recovered from mice immunized with the E1/sE2F442NYT-mRNA preparation displayed a minimum of a 2-fold enhancement in neutralization efficacy on HCVpp compared to the E1/sE2-immunized mice (Fig. 6A). To further evaluate protection generated by immunization with E1/sE2 mRNA-LNP variants, we challenged mice with a surrogate recombinant vaccinia virus expressing HCV E1-E2-NS2aa134-966 from HCV genotype 1a (clone H77). Immunized mice were sacrificed 4 days after challenge infection. Interestingly, vehicle control or unmodified E1/sE2 mRNA vaccinated mice failed to show a statistically different level in virus recovery from the ovary homogenate by plaque assay (Fig. 6B). In contrast, the E1/sE2F442NYT mRNA vaccine displayed a significant reduction (≥4 log) in virus recovery from the ovaries of challenged mice. Two of five mice showed minimal virus recovery, while the remaining three displayed an undetected virus titer. We repeated the challenge study with an additional group of mice (five in each group) under identical immunization, challenge, and experimental protocols. All E1/sE2F442NYT mRNA-vaccinated mice displayed an undetected virus titer in the ovary after challenge infection. On the other hand, vehicle control or unmodified E1/sE2 mRNA-vaccinated mice failed to show a statistically different level in virus recovery from by plaque assay. These data indicate that E1/sE2F442NYT mRNA-LNP vaccine preparation induced a significantly improved protective immune response in immunized animals.
FIG 6.
Immunized mouse sera induce HCV pseudotype neutralization and protection from HCV vaccinia challenge. (A) Neutralization of lentivirus-derived HCV pseudotype particles (HCVpp) as a surrogate virus was carried out with individual mouse sera at serial dilutions (1/400 to 1/3,200) from the immunized animals (five in each group). (B) Immunized female BALB/c mice were examined for protection from challenge infection by recombinant vaccinia virus expressing HCV E1-E2-NS2aa134-966. Mouse ovaries were collected after challenge infection, homogenized in medium, freeze-thawed three times, and centrifuged to pellet cell debris. Clear supernatant was serially 10-fold diluted and plaque assayed for virus recovery on BSC40 cells. Crystal violet-stained plaques were counted, and the results are expressed as PFU/mL. The significance level is indicated (*, P < 0.05; ***, P < 0.001; ns, not significant).
DISCUSSION
HCV E2 induces immune regulatory cytokines and interrupts the function of immunity-related cells by binding with CD81 (7–9, 24). Here, we examined whether a modified sE2 antigen inhibiting CD81 binding induces an improved immunogenic profile for vaccine development. We have addressed three novel aspects in this area: (i) modulation of E2 reduces CD81 binding and alters proinflammatory cytokine response in human PBMCs and the use of mRNA-LNP vaccine platform (ii) induces a strong immune response in a mouse model and (iii) potentiates clearance of HCV vaccinia challenge infection. We conducted in vitro studies using human PBMCs for comparison of response to wild-type and modified sE2 antigens and subsequently included a mouse HCV vaccinia challenge model to examine immune parameters already observed with human PBMCs. Enhanced CD4+ T helper cell-associated cytokine expression was observed in human PBMCs and in mice immunized with mRNA-LNP vaccine expressing E1 and modified sE2 (E1/sE2F44NYT). Protective immunity against HCV in mice was verified by challenge infection with a surrogate recombinant vaccinia virus. Immunity in mice is likely mediated by cytokines associated with CD4+ and CD8+ T cell activation. Interestingly, modified sE2F442NYT appeared as a potent inducer of cellular immune responses compared to wild-type sE2. The cytokine profile of HCV-specific T cells suggested an induction of a Th1 cytokine profile that may help in viral clearance.
We observed an alteration of the cytokine response and improved protection to challenge HCV vaccinia virus in a mouse model. The use of modified sE2 resulted in a 2- to 4-fold-higher neutralizing activity, and this approach could be used for mutations and combinations with a mRNA-LNP vaccine platform. We need to better understand the durability of neutralizing antibodies and the contributions of B and T cells in the clearance of challenge HCV vaccinia virus in immunized mice. We included E1 in our study as an additional antigen for additive antiviral immune response, and this will require further study to determine whether it provides a protective advantage for the inhibition of an HCV vaccinia challenge infection.
Macrophages play a crucial role in antigen presentation and in the interaction between innate and adaptive immunity. M1 macrophages promote a Th1 response and possess antiviral activity, while M2 macrophages are involved in the promotion of a Th2 response and immune tolerance (25). In a previous safety and immunogenicity vaccine study in humans, purified HCV E1/E2 envelope glycoproteins administered with MF59 induced a weak immune response (26). MF59 was used as an adjuvant to facilitate a Th1 cytokine profile and the induction of CD4+ memory and cytotoxic T-lymphocyte responses. However, the vaccinated volunteers exhibited induction of IL-10, with only a minimal increase in IL-12 expression. Our in vitro studies suggest that HCV E2 induces IL-10 in human PBMC-derived macrophages, lowers IL-12 production (6), and promotes M2 phenotype polarization of macrophages.
Glycosylation has a role in adaptive immune activation (27). Strategic glycosylation of antigens is exploited for the development of more efficacious prophylactic and therapeutic vaccines (28). Preservation of all the characterized neutralizing epitopes may not be possible in antigen selection for potential vaccine development. The E2 front layer constitutes most of the CD81 binding sites, remarkably flexible, and the amino acid residues 424 to 453 may be highly immunogenic, skewing the immune response toward nonneutralizing epitopes (29). E2 is densely glycosylated, and the CD81 receptor-binding site is not obstructed by any of the 11 N-linked glycans (26). Removal of N-linked glycosylation sites appears to alter the antigenic properties of HCV E2 (29). Our results from the present study indicate that interruption of CD81 binding and introduction of a potential N-linked glycosylation site on the modified sE2 antigen, sE2F442NYT, results in an improved T cell response that switches from the generation of anti-inflammatory cytokines to the generation of proinflammatory cytokines compared to a single amino acid substitution in sE2L427Y. Further examination of this altered site in future will verify the glycosylation status of the modified E2 and gain in vaccine antigen selection. The modified sE2F442NYT helped in the generation of proinflammatory cytokines (IFN-γ, IL-12, IL-6, and TNF-α), while sE2L427Y did not to a similar extent.
IL-10 is a pleiotropic master regulatory cytokine produced by various immune cells, primarily macrophages, DCs, and T-helper cells. IL-10 critically associates with the persistence of viruses by suppressing cell-mediated immunity and represents a major immunosuppressive cytokine. The role of IL-10 is to limit the extent of the activation of both innate and adaptive immune cells to maintain a homeostatic state. IL-10 regulates the growth and/or differentiation of B cells, NK cells, cytotoxic and helper T cells, mast cells, granulocytes, DCs, keratinocytes, and endothelial cells (30). On the other hand, IL-12 orchestrates resistance against infectious diseases through macrophage activation and the induction of IFN-γ and may help in eliminating intracellular pathogens (31). In chronically HCV-infected patients, T cells are inefficient and defective in producing IFN-γ (32). IL-12 and IFN-γ work concomitantly in the development of a Th1 immune response against viral infections. Exposure of macrophage to IL-10 suppresses IFN-γ-induced genes (17). An early protective Th1 response favoring IFN-γ production in the presence of IL-12 would be beneficial for a host to prevent virus infection. Resistance of mice to surrogate recombinant VV E1-E2-NS2aa134-966 challenge infection after immunization using a nucleoside-modified mRNA-LNP vaccine encoding E1/sE2F442NYT supports the potential use of modified E2 in an HCV vaccine.
HCV induces IL-10 in monocyte-derived DCs and inhibits DC-mediated antigen-specific T cell activation. IL-12 activates NK cells and induces the differentiation of naive CD4+ T lymphocytes to become IFN-γ-producing Th1 effectors in cell-mediated immune responses to intracellular pathogens (33). IFN-γ priming is a positive-feedback mechanism for more robust IL-12 production in certain immune responses. Th1 lymphocytes are initially activated by antigen-presenting-cell-derived IL-12 upon pathogen infection. Individuals who spontaneously clear HCV display broad CD4+ T cell responses, stronger T cell proliferation, and higher IL-2, IFN-γ, and TNF-α production than individuals who develop chronic infection dominated by Tregs and IL-10 production (34, 35). Strong HCV-specific CD8+ T cell responses are generated, and HCV-specific memory T cells persist after recovery from infection (36–39).
An effective vaccine is a major unmet need to achieve global elimination of HCV, and there is potential for HCV vaccine development (3, 40). A recent report (5) suggested that an HCV T cell-based vaccine regimen encoding nonstructural (NS) proteins generated specific T cell responses and lowered the peak HCV RNA level but did not prevent chronic infection. We have shown previously that cells endogenously expressing HCV proteins perturb HLA-DR cell surface expression (16). HCV E2 induces immune regulatory cytokine IL-10 and CD163 protein expression in primary macrophages (6) and inhibits the function of T, B, and NK cells through binding with CD81 (7, 8, 24, 41). HCV-specific CD4+ T cell immune deficiency may act as a primary cause of CD8+ T cell functional exhaustion (10, 37).
HCV exhibits extensive genetic diversity. An optimal HCV vaccine probably needs broadly cross-reactive cellular immune responses and cross-neutralizing antibodies. Priming of CD4+ and CD8+ T cells specific for HCV modified sE2 antigens may not induce an immune response to all the antigenic sites or epitopes, but our approach will likely increase the efficacy of the overall vaccine significantly to confer HCV immunity. The candidate HCV sE2 mutant mRNA-LNP vaccine elicits innate, humoral, and T cell responses to target T and B cell determinants. Further studies are necessary to characterize CD4+ and CD8+ T cell responses and the duration of immunity induced by modified HCV sE2 antigen and understanding the beneficial role of added E1 antigen, although sE2 is unlikely to form a heterodimer with the E1 protein under these conditions. The use of other animal models developed in the future will provide further help for HCV vaccine evaluation. The results from our study will considerably advance highly effective and robust vaccine development against persistent hepatotropic HCV across genotypes by using the mRNA-LNP vaccine platform.
MATERIALS AND METHODS
Cells.
Human embryonic kidney 293T (HEK293T), human monocyte-derived primary macrophages, and human-derived CD4+T cells were used. HEK293T cells were maintained in Dulbecco modified Eagle medium containing penicillin-streptomycin, 10% fetal calf serum (FCS), 1% l-glutamine, and 1% nonessential amino acids. Primary macrophages and primary CD4+T cells were maintained in RPMI containing penicillin-streptomycin supplemented with 10% FCS and 1% l-glutamine.
Site-specific mutagenesis of sE2.
HCV (H77c genotype 1a) E2 sequence corresponding to 383 to 661 amino acids cloned into the pcDNA 3.1 vector (a gift from Heidi E. Drummer) was used as a template for the generation of sE2 mutants by a QuikChange Lightning site-directed mutagenesis kit (Agilent Technology) following the supplier’s procedure. E2 was expressed as C-terminal truncations in mammalian cells, since soluble and correctly folded E2 can be obtained by deleting the transmembrane domain (42).
The sE2 single mutant (L427Y) was generated by replacing a nonpolar leucine (L) amino acid residue at position 427 with a polar amino acid tyrosine (Y). The sE2 double mutant (F442NYT) was generated on the same E2 sequence by replacing two amino acids, as indicated. One replacement was done at position 442 with a nonpolar amino acid phenylalanine (F) to a polar amino acid asparagine (N), and another replacement occurred at position 444 of a polar amino acid glutamine (Q) with a polar amino acid threonine (T). The alterations at amino acid positions 442 and 444 (FYQ→NYT) included a N-linked glycosylation site based on an earlier publication (15). The L427Y forward primer (5′-GCACATCAATAGCACGGCCTATAACTGCAATGAAAGCCTTAA-3′) and reverse primer (5′-TTAAGGCTTTCATTGCAGTTATAGGCCGTGCTATTGATGTGC-3′) and the F442NYT forward primer (5′-ACCGGCTGGTTAGCAGGGCTCAACTATACGCACAAAT-3′) and reverse primer (5′-ATTTGTGCGTATAGTTGAGCCCTGCTAACCAGCCGGT-3′) were used for site-specific mutagenesis. The sequence changes in constructs were confirmed by plasmid isolation and DNA sequencing.
Cell transfection, protein expression, and purification.
The plasmid DNA isolated from clones were transfected into HEK293T cells using Lipofectamine 3000 transfection reagent (Thermo Fisher). Culture supernatant was harvested after 48 h, passed through a 0.45-μm-pore size filter (Nalgene), and stored at −20°C until purification of the recombinant proteins containing a hexahistidine tag. Proteins were purified using a nickel-NTA-affinity column (HisPur Ni-NTA spin columns; Thermo Fisher) and verified by Coomassie blue staining and Western blot analysis with anti-His antibody (Sino Biological).
E2-CD81 binding assay.
Recombinant human CD81 protein (R&D Systems) was coated (4 μg/mL) on an ELISA plate, and 100 or 200 μL of sE2 or modulated HEK293T-transfected culture supernatant or 1.25, 2.5, and 5.0 μg/mL of purified protein (in triplicates) was added. Horseradish peroxidase-conjugated anti-His antibody (Sino Biological), followed by TMB substrate, was added, and the reaction read at 450 nm in a microplate reader.
Stimulation of PBMCs.
PBMCs were isolated from blood by using Ficoll-Paque Plus (GE Healthcare) density gradient centrifugation. Cells (1 × 106)/well were plated, and nonadherent cells were removed and incubated for the maturation of monocytes to macrophages. On day 5, macrophages were stimulated with sE2 (2.5 μg/mL) or modified protein for 24 h at 37°C. Clarified cell culture supernatants and lysed adherent cells were stored frozen.
Cytokine quantification.
Secretory cytokines present in the cell culture supernatant of macrophages exposed to HCV sE2, sE2L427Y, or sE2F442NYT after 24 h were quantified by using commercially available paired antibodies for an IL-10, IL-12, IL-6, IFN-γ, and TNF-α cytokine kit (Sino Biological). IL-4 was quantified using a separate kit (Thermo Fisher). Culture supernatants from activated CD4+T cells exposed to HCV sE2 or sE2F442NYT were similarly quantified after 72 h for IFN-γ and IL-4. Mouse cytokine levels in vaccinated animals (IL-4, IL-10, IL-12, and IFN-γ) were measured by ELISA (Invitrogen or R&D Systems).
Macrophage activation marker analysis.
Monocyte-derived macrophages in culture were stimulated with 2.5 μg/mL sE2 or modified sE2 protein for 24 h. Culture supernatants were removed, and cells were collected by dislodging them with 5 mM EDTA, followed by centrifugation and suspension for flow cytometry with CD80-FITC, CD86-PE, CD40-PE, or HLA-DR APC (BioLegend).
Macrophage/T cell coculture and Th1 and Th2 marker analyses.
PBMCs from healthy donors were used for CD4+T cell isolation (Miltenyi Biotec) and treated with recombinant IL-2. Matured monocyte-derived macrophages were seeded in a 48-well plate (1 × 105/well) and stimulated with sE2 (2.5 μg/mL) or modified sE2 for 24 h. Live T cells (2.5 × 104) and macrophages (1 × 105) were cocultured for 4 days. Cells were harvested on day 5 and stained with CD4-FITC, CXCR3-PE, and CCR4-APC antibodies (BioLegend) for FACS analysis.
CD4+T cell proliferation.
PBMCs from healthy donors were used for CD4+T cell isolation (Miltenyi Biotec) and stained with 3 μM CFSE (Invitrogen). Washed cells were activated by using 5 μg/mL human anti-CD3 antibody and 1 μg/mL human anti-CD28 antibody (BioLegend), followed by incubation for 3 days in the presence or absence of 2.5 μg/mL sE2 or sE2F442NQT for flow analysis.
T cell activation marker analysis.
Isolated CD4+T cells from healthy donors were incubated with sE2 or sE2F442NYT in the presence of 5 μg/mL human anti-CD3 antibody and 1 μg/mL human anti-CD28 antibody (BioLegend) for 2 h. Cells were harvested and stained with CD69-FITC or CD25-PE (BioLegend) for FACS analysis.
Generation of mRNA-LNP vaccines encoding the luciferase and HCV protein.
Codon-optimized luciferase HCV E1193-351 and HCV E2386-660 specific sequences with or without mutations at residues 442 and 444 were synthesized into mRNAs, purified and loaded as LNP candidate vaccines formulated for immunization of mice in combination for this study. mRNA-loaded LNPs were formulated using a total lipid concentration of 40 mM, as previously described (43). The ethanolic lipid mixture comprising ionizable cationic lipid, phosphatidylcholine, cholesterol, and polyethylene glycol-lipid was rapidly mixed with an aqueous solution containing cellulose-purified N1-mΨ in vitro-transcribed mRNAs. The LNP formulation used in this study is proprietary to Acuitas Therapeutics (US patent 10,221,127). RNA-loaded particles were characterized for their size, surface charge, encapsulation efficiency, and endotoxin content and subsequently stored at −80°C at an RNA concentration of 1 μg/μL (in the case of loaded particles) and a total lipid concentration of 30 μg/μL (both loaded and empty particles). The mean hydrodynamic diameter of mRNA-LNPs was ~80 nm, with a polydispersity index of 0.02 to 0.06 and an encapsulation efficiency of ~95%. Two or three batches from each mRNA-LNP formulations were used in these studies.
mRNA-LNP vaccine immunization and challenge infection using VV/HCVaa134-966.
BALB/c mice (Jackson Lab) were divided into groups (five mice per group) and immunized intramuscularly in the hind leg with mRNA-LNP candidate vaccine as E1/sE2 or E1/sE2F442NTY (10 μg of vaccine antigen) twice at 2-week intervals. Test bleeds (3 days before immunization and 3 days before challenge infection) from mice were analyzed. The HCV vaccinia challenge as a surrogate model is helpful to suggest induction of protective immunity (44). Immunized mice were challenged intraperitoneally with live recombinant vaccinia virus expressing HCVE1-E2-NS2134-966 (45) and sacrificed 4 days after challenge infection for collection of the ovaries for further analysis. The recombinant vaccinia virus dose was chosen for mice inoculation used previously for other immune protection-related studies (46–48). Ovaries were homogenized in 300 μL of medium by a motor-operated disposable sterile homogenizer, freeze-thawed three times, and centrifuged to pellet cell debris. Clear supernatant was serially 10-fold diluted and plaque assayed to determine the virus titer on a BSC-40 cell monolayer. Plaques were stained after 3 days with 1% crystal violet.
Statistical analysis.
A Student two-tailed paired t test or an unpaired t test was used to determine the difference within the groups studied. One- or two-way analysis of variance (ANOVA) was used to compare between the groups. All the data are expressed as means ± the standard errors of the mean, and a P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Heidi Drummer, Burnet Institute, Australia, for providing the HCV sE2 clone and Michael Houghton, University of Alberta, Canada, for providing recombinant VVHCVE1-E2-NS2aa134-966. We thank Jenni Franey from Comparative Medicine, Saint Louis University, for technical help in the vaccinia challenge experiments.
This study was supported by National Institutes of Health grants R01-DK122401 (R.R.) and R01-AI124429 (D.W.).
Conceptualization—R.R. conceptualized the research plan of the study, and D.W. conceptualized the use of mRNA-LNP vaccine approach; methodology—V.V., T.P., K.M., M.G.-A., and E.K.R.; investigation—V.V., T.P., K.M., M.G.-A., E.K.R., D.W., and R.R.; funding acquisition—R.R. and D.W.; writing—V.V., T.P., K.M., M.G.-A., E.K.R., and R.R.; and review and editing—R.R. and D.W.
Footnotes
[This article was published on 25 May 2022 with minor errors in Materials and Methods. The text was updated in the current version, posted on 22 June 2022.]
Contributor Information
Ranjit Ray, Email: ranjit.ray@health.slu.edu.
J.-H. James Ou, University of Southern California
REFERENCES
- 1.Hamdane N, Juhling F, Crouchet E, El Saghire H, Thumann C, Oudot MA, Bandiera S, Saviano A, Ponsolles C, Roca Suarez AA, Li S, Fujiwara N, Ono A, Davidson I, Bardeesy N, Schmidl C, Bock C, Schuster C, Lupberger J, Habersetzer F, Doffoel M, Piardi T, Sommacale D, Imamura M, Uchida T, Ohdan H, Aikata H, Chayama K, Boldanova T, Pessaux P, Fuchs BC, Hoshida Y, Zeisel MB, Duong FHT, Baumert TF. 2019. HCV-induced epigenetic changes associated with liver cancer risk persist after sustained virologic response. Gastroenterology 156:2313–2329. 10.1053/j.gastro.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thimme R. 2021. T cell immunity to hepatitis C virus: lessons for a prophylactic vaccine. J Hepatol 74:220–229. 10.1016/j.jhep.2020.09.022. [DOI] [PubMed] [Google Scholar]
- 3.Houghton M. 2011. Prospects for prophylactic and therapeutic vaccines against the hepatitis C viruses. Immunol Rev 239:99–108. 10.1111/j.1600-065X.2010.00977.x. [DOI] [PubMed] [Google Scholar]
- 4.Ray R, Meyer K, Banerjee A, Basu A, Coates S, Abrignani S, Houghton M, Frey SE, Belshe RB. 2010. Characterization of antibodies induced by vaccination with hepatitis C virus envelope glycoproteins. J Infect Dis 202:862–866. 10.1086/655902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Page K, Melia MT, Veenhuis RT, Winter M, Rousseau KE, Massaccesi G, Osburn WO, Forman M, Thomas E, Thornton K, Wagner K, Vassilev V, Lin L, Lum PJ, Giudice LC, Stein E, Asher A, Chang S, Gorman R, Ghany MG, Liang TJ, Wierzbicki MR, Scarselli E, Nicosia A, Folgori A, Capone S, Cox AL. 2021. Randomized trial of a vaccine regimen to prevent chronic HCV infection. N Engl J Med 384:541–549. 10.1056/NEJMoa2023345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon YC, Meyer K, Peng G, Chatterjee S, Hoft DF, Ray R. 2019. Hepatitis C virus E2 envelope glycoprotein induces an immunoregulatory phenotype in macrophages. Hepatology 69:1873–1884. 10.1002/hep.29843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosa D, Saletti G, De Gregorio E, Zorat F, Comar C, D’Oro U, Nuti S, Houghton M, Barnaba V, Pozzato G, Abrignani S. 2005. Activation of naive B lymphocytes via CD81, a pathogenetic mechanism for hepatitis C virus-associated B lymphocyte disorders. Proc Natl Acad Sci USA 102:18544–18549. 10.1073/pnas.0509402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crotta S, Ronconi V, Ulivieri C, Baldari CT, Valiante NM, Valiente NM, Abrignani S, Wack A. 2006. Cytoskeleton rearrangement induced by tetraspanin engagement modulates the activation of T and NK cells. Eur J Immunol 36:919–929. 10.1002/eji.200535527. [DOI] [PubMed] [Google Scholar]
- 9.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 10.Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, Murthy KK, Rice CM, Walker CM. 2003. HCV persistence and immune evasion in the absence of memory T cell help. Science 302:659–662. 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 11.Kumar A, Pandey R, Yadav IS, Bharadwaj M. 2018. Structural and epitope analysis (T- and B-cell epitopes) of hepatitis C virus (HCV) glycoproteins: an in silico approach. J Clin Exp Hepatol 8:352–361. 10.1016/j.jceh.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vijayamahantesh V, Patra T, Meyer K, Mohamed-Gabriel A, Reagan E, Weissman D, Ray R. 2022. Modified E2 glycoprotein of hepatitis C virus enhances pro-inflammatory cytokines and protective immune response in mice. bioRxiv. https://www.biorxiv.org/content/10.1101/2022.02.07.479452v1. [DOI] [PMC free article] [PubMed]
- 13.Petracca R, Falugi F, Galli G, Norais N, Rosa D, Campagnoli S, Burgio V, Di Stasio E, Giardina B, Houghton M, Abrignani S, Grandi G. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J Virol 74:4824–4830. 10.1128/jvi.74.10.4824-4830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pierce BG, Keck ZY, Wang R, Lau P, Garagusi K, Elkholy K, Toth EA, Urbanowicz RA, Guest JD, Agnihotri P, Kerzic MC, Marin A, Andrianov AK, Ball JK, Mariuzza RA, Fuerst TR, Foung SKH. 2020. Structure-based design of hepatitis C virus E2 glycoprotein improves serum binding and cross-neutralization. J Virol 94:e00704-20. 10.1128/JVI.00704-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, Wilson IA, Law M. 2013. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342:1090–1094. 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito K, Ait-Goughoulte M, Truscott SM, Meyer K, Blazevic A, Abate G, Ray RB, Hoft DF, Ray R. 2008. Hepatitis C virus inhibits cell surface expression of HLA-DR, prevents dendritic cell maturation, and induces interleukin-10 production. J Virol 82:3320–3328. 10.1128/JVI.02547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ray R. 2011. Progress toward development of a hepatitis C vaccine with broad shoulders. Sci Transl Med 3:94ps33. [DOI] [PubMed] [Google Scholar]
- 18.Kane MM, Mosser DM. 2001. The role of IL-10 in promoting disease progression in leishmaniasis. J Immunol 166:1141–1147. 10.4049/jimmunol.166.2.1141. [DOI] [PubMed] [Google Scholar]
- 19.Bertani FR, Mozetic P, Fioramonti M, Iuliani M, Ribelli G, Pantano F, Santini D, Tonini G, Trombetta M, Businaro L, Selci S, Rainer A. 2017. Classification of M1/M2-polarized human macrophages by label-free hyperspectral reflectance confocal microscopy and multivariate analysis. Sci Rep 7:8965. 10.1038/s41598-017-08121-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Smith W, Hao D, He B, Kong L. 2019. M1 and M2 macrophage polarization and potentially therapeutic naturally occurring compounds. Int Immunopharmacol 70:459–466. 10.1016/j.intimp.2019.02.050. [DOI] [PubMed] [Google Scholar]
- 21.Dong C, Flavell RA. 2000. Cell fate decision: T-helper 1 and 2 subsets in immune responses. Arthritis Res 2:179–188. 10.1186/ar85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorfman DM, Shahsafaei A. 2002. CD69 expression correlates with expression of other markers of Th1 T cell differentiation in peripheral T cell lymphomas. Hum Pathol 33:330–334. 10.1053/hupa.2002.32215. [DOI] [PubMed] [Google Scholar]
- 23.Schulze Zur Wiesch J, Ciuffreda D, Lewis-Ximenez L, Kasprowicz V, Nolan BE, Streeck H, Aneja J, Reyor LL, Allen TM, Lohse AW, McGovern B, Chung RT, Kwok WW, Kim AY, Lauer GM. 2012. Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J Exp Med 209:61–75. 10.1084/jem.20100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crotta S, Stilla A, Wack A, D’Andrea A, Nuti S, D’Oro U, Mosca M, Filliponi F, Brunetto RM, Bonino F, Abrignani S, Valiante NM. 2002. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J Exp Med 195:35–41. 10.1084/jem.20011124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantovani A, Sica A. 2010. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol 22:231–237. 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Frey SE, Houghton M, Coates S, Abrignani S, Chien D, Rosa D, Pileri P, Ray R, Di Bisceglie AM, Rinella P, Hill H, Wolff MC, Schultze V, Han JH, Scharschmidt B, Belshe RB. 2010. Safety and immunogenicity of HCV E1E2 vaccine adjuvanted with MF59 administered to healthy adults. Vaccine 28:6367–6373. 10.1016/j.vaccine.2010.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderluh M, Berti F, Bzducha-Wróbel A, Chiodo F, Colombo C, Compostella F, Durlik K, Ferhati X, Holmdahl R, Jovanovic D, Kaca W, Lay L, Marinovic-Cincovic M, Marradi M, Ozil M, Polito L, Reina-Martin JJ, Reis CA, Sackstein R, Silipo A, Švajger U, Vaněk O, Yamamoto F, Richichi B, van Vliet SJ. 2021. Emerging glyco-based strategies to steer immune responses. FEBS J 288:4746–4772. 10.1111/febs.15830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avci FY, Li X, Tsuji M, Kasper DL. 2011. A mechanism for glycoconjugate vaccine activation of the adaptive immune system and its implications for vaccine design. Nat Med 17:1602–1609. 10.1038/nm.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prentoe J, Velazquez-Moctezuma R, Augestad EH, Galli A, Wang R, Law M, Alter H, Bukh J. 2019. Hypervariable region 1 and N-linked glycans of hepatitis C regulate virion neutralization by modulating envelope conformations. Proc Natl Acad Sci USA 116:10039–10047. 10.1073/pnas.1822002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. 2001. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol 19:683–765. 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 31.Murray SM, Linial ML. 2006. Foamy virus infection in primates. J Med Primatol 35:225–235. 10.1111/j.1600-0684.2006.00171.x. [DOI] [PubMed] [Google Scholar]
- 32.Semmo N, Klenerman P. 2007. CD4+ T cell responses in hepatitis C virus infection. World J Gastroenterol 13:4831–4838. 10.3748/wjg.v13.i36.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma X, Yan W, Zheng H, Du Q, Zhang L, Ban Y, Li N, Wei F. 2015. Regulation of IL-10 and IL-12 production and function in macrophages and dendritic cells. F1000Res 4:1465. 10.12688/f1000research.7010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urbani S, Amadei B, Fisicaro P, Tola D, Orlandini A, Sacchelli L, Mori C, Missale G, Ferrari C. 2006. Outcome of acute hepatitis C is related to virus-specific CD4 function and maturation of antiviral memory CD8 responses. Hepatology 44:126–139. 10.1002/hep.21242. [DOI] [PubMed] [Google Scholar]
- 35.Smyk-Pearson S, Tester IA, Klarquist J, Palmer BE, Pawlotsky JM, Golden-Mason L, Rosen HR. 2008. Spontaneous recovery in acute human hepatitis C virus infection: functional T-cell thresholds and relative importance of CD4 help. J Virol 82:1827–1837. 10.1128/JVI.01581-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P, Robbins G, Phillips R, Klenerman P, Walker BD. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med 191:1499–1512. 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lauer GM. 2013. Immune responses to hepatitis C virus (HCV) infection and the prospects for an effective HCV vaccine or immunotherapies. J Infect Dis 207(Suppl 1):S7–S12. 10.1093/infdis/jis762. [DOI] [PubMed] [Google Scholar]
- 38.Takaki A, Wiese M, Maertens G, Depla E, Seifert U, Liebetrau A, Miller JL, Manns MP, Rehermann B. 2000. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat Med 6:578–582. 10.1038/75063. [DOI] [PubMed] [Google Scholar]
- 39.Shata MT, Anthony DD, Carlson NL, Andrus L, Brotman B, Tricoche N, McCormack P, Prince A. 2002. Characterization of the immune response against hepatitis C infection in recovered, and chronically infected chimpanzees. J Viral Hepat 9:400–410. 10.1046/j.1365-2893.2002.00373.x. [DOI] [PubMed] [Google Scholar]
- 40.Cox AL. 2020. Challenges and promise of a hepatitis C virus vaccine. Cold Spring Harb Perspect Med 10:a036947. 10.1101/cshperspect.a036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wack A, Soldaini E, Tseng C, Nuti S, Klimpel G, Abrignani S. 2001. Binding of the hepatitis C virus envelope protein E2 to CD81 provides a costimulatory signal for human T cells. Eur J Immunol 31:166–175. . [DOI] [PubMed] [Google Scholar]
- 42.McCaffrey K, Boo I, Owczarek CM, Hardy MP, Perugini MA, Fabri L, Scotney P, Poumbourios P, Drummer HE. 2017. An optimized hepatitis C virus E2 glycoprotein core adopts a functional homodimer that efficiently blocks virus entry. J Virol 91:e01668-16. 10.1128/JVI.01668-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alameh MG, Tombacz I, Bettini E, Lederer K, Sittplangkoon C, Wilmore JR, Gaudette BT, Soliman OY, Pine M, Hicks P, Manzoni TB, Knox JJ, Johnson JL, Laczko D, Muramatsu H, Davis B, Meng W, Rosenfeld AM, Strohmeier S, Lin PJC, Mui BL, Tam YK, Kariko K, Jacquet A, Krammer F, Bates P, Cancro MP, Weissman D, Luning Prak ET, Allman D, Locci M, Pardi N. 2021. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 54:2877–2892. 10.1016/j.immuni.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olivera S, Perez A, Falcon V, Urquiza D, Pichardo D, Martinez-Donato G. 2020. Protective cellular immune response against hepatitis C virus elicited by chimeric protein formulations in BALB/c mice. Arch Virol 165:593–607. 10.1007/s00705-019-04464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray R, Khanna A, Lagging LM, Meyer K, Choo QL, Ralston R, Houghton M, Becherer PR. 1994. Peptide immunogen mimicry of putative E1 glycoprotein-specific epitopes in hepatitis C virus. J Virol 68:4420–4426. 10.1128/JVI.68.7.4420-4426.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murata K, Lechmann M, Qiao M, Gunji T, Alter HJ, Liang TJ. 2003. Immunization with hepatitis C virus-like particles protects mice from recombinant hepatitis C virus-vaccinia infection. Proc Natl Acad Sci USA 100:6753–6758. 10.1073/pnas.1131929100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alvarez-Lajonchere L, Gonzalez M, Alvarez-Obregon JC, Guerra I, Vina A, Acosta-Rivero N, Musacchio A, Morales J, Duenas-Carrera S. 2006. Hepatitis C virus (HCV) core protein enhances the immunogenicity of a co-delivered DNA vaccine encoding HCV structural antigens in mice. Biotechnol Appl Biochem 44:9–17. 10.1042/BA20050202. [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Donato G, Piniella B, Aguilar D, Olivera S, Perez A, Castanedo Y, Alvarez-Lajonchere L, Duenas-Carrera S, Lee JW, Burr N, Gonzalez-Miro M, Rehm BH. 2016. Protective T cell and antibody immune responses against hepatitis C virus achieved using a biopolyester-bead-based vaccine delivery system. Clin Vaccine Immunol 23:370–378. 10.1128/CVI.00687-15. [DOI] [PMC free article] [PubMed] [Google Scholar]