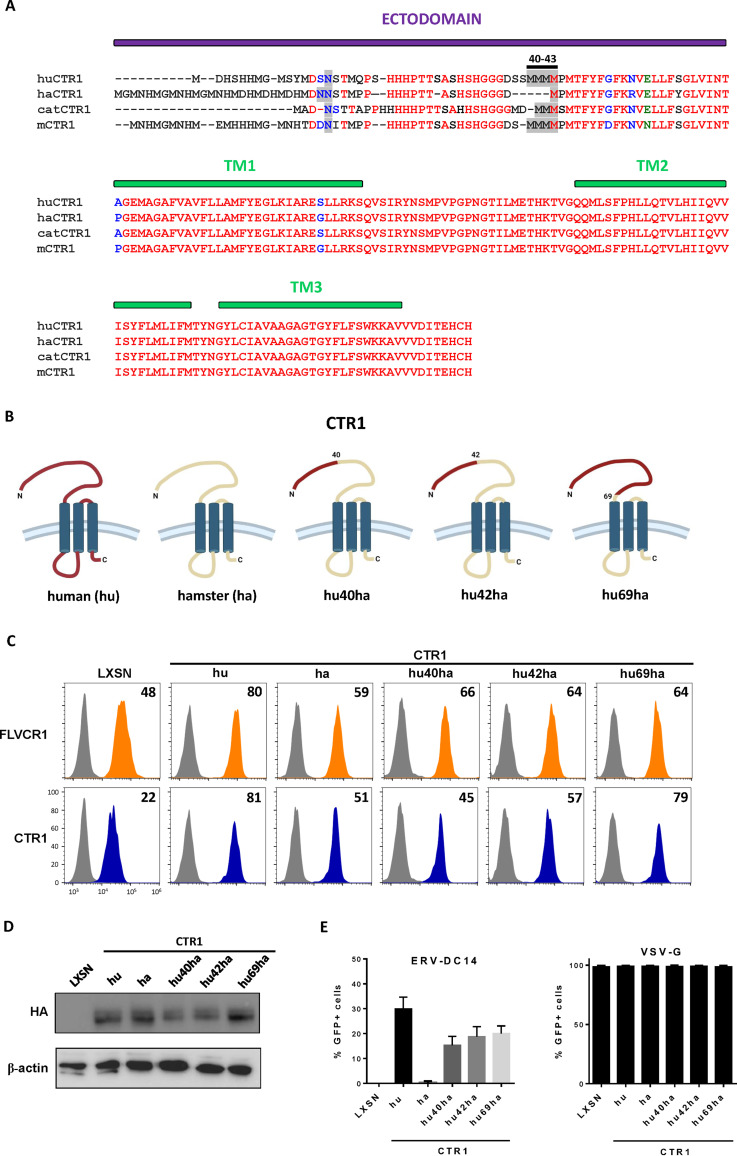
FIG 5.
Functional analysis of human/hamster CTR1 chimeras. (A) Amino acid sequences of human, hamster, cat, and mouse CTR1 were aligned using ClustalW. The ectodomain and the 3 transmembranes (TM) are indicated in purple and green boxes, respectively. The N-linked glycosylation sites and the quadruplet of methionines at positions 40 to 43 of the human sequence are highlighted in gray. Amino acid color code is as follows: red, identical; green, strongly similar; blue, weakly similar; black, different. Deletions are indicated with dashes. (B) Schematic representations of chimeric constructs between human and hamster CTR1. (C) CHO cells stably transduced with LXSN retroviral vector either empty or carrying the CTR1 cDNA from human (hu), hamster (ha), or chimeras (hu40ha, hu42ha, and hu69ha) were evaluated for CTR1 or FLVCR1 cell surface expression by flow cytometry using the RBD ligands. Numbers (× 103) indicate the delta mean fluorescence intensity of a representative experiment (n = 3). (D) Representative immunoblot of HA-tagged CTR1 in cell lysates from panel C. (E) Cells from panel C were evaluated for their sensitivity to infection by EGFP retroviral vectors pseudotyped with either the ERV-DC14-specific Env or the VSV-G protein. Data are means ± SEM from n = 3 experiments.