Abstract
Munc18-1 forms a template to organize assembly of the neuronal SNARE complex that triggers neurotransmitter release, binding first to a closed conformation of syntaxin-1 where its amino-terminal region interacts with the SNARE motif, and later binding to synaptobrevin. However, the mechanism of SNARE complex assembly remains unclear. Here, we report two cryo-EM structures of Munc18-1 bound to cross-linked syntaxin-1 and synaptobrevin. The structures allow visualization of how syntaxin-1 opens and reveal how part of the syntaxin-1 amino-terminal region can help nucleate interactions between the amino termini of the syntaxin-1 and synaptobrevin SNARE motifs, while their carboxyl termini bind to distal sites of Munc18-1. These observations, together with mutagenesis, SNARE complex assembly experiments, and fusion assays with reconstituted proteoliposomes, support a model whereby these interactions are critical to initiate SNARE complex assembly and multiple energy barriers enable diverse mechanisms for exquisite regulation of neurotransmitter release.
Cryo-EM structures reveal key insights into the molecular mechanism of neuronal SNARE complex assembly guided by Munc18-1.
INTRODUCTION
Release of neurotransmitters by Ca2+-evoked synaptic vesicle exocytosis is mediated by the vesicle soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) synaptobrevin and the plasma membrane SNAREs syntaxin-1 and SNAP-25, which form a tight complex (1) through their ~65-residue SNARE motifs. The SNARE complex consists of a parallel four-helix bundle (2, 3) and brings the vesicle and plasma membranes together (4), which is crucial for membrane fusion [reviewed in (5)]. In syntaxin-1, the SNARE motif is preceded by a 190-residue N-terminal region (Fig. 1A) containing a three-helix bundle domain (the Habc domain) that binds intramolecularly to the SNARE motif, forming a “closed” conformation that precludes SNARE complex assembly (6–8). The SNARE complex is disassembled by N-ethylmaleimide–sensitive factor (NSF) and soluble NSF attachment proteins (SNAPs; no relation to SNAP-25) (1), whereas SNARE complex assembly is orchestrated through an NSF-SNAP–resistant pathway by Munc18-1 and Munc13 (9, 10). This pathway starts with Munc18-1 bound tightly to closed syntaxin-1 (7, 8) and is activated by Munc13 when it helps to open syntaxin-1 (11, 12), while it bridges the vesicle and plasma membranes (13, 14), enabling a wide variety of Munc13-dependent presynaptic plasticity processes that underlie multiple forms of information processing in the brain (15–17). The importance of this pathway was highlighted by the total abrogation of neurotransmitter release observed in the absence of Munc18-1 or Munc13s (18–21). Moreover, an L165A-E166A mutation that opens syntaxin-1 (LE mutation) (7) partially rescues the impairments in release caused by deletion of diverse proteins in Caenorhabditis elegans, including the Munc13 homolog Unc-13 (15, 22, 23), and a P335A mutation in Munc18-1 that leads to a gain of function (24–26) also rescues release partially in unc-13 nulls (16). Thus, elucidating the mechanism of SNARE complex assembly not only is critical to understand brain function but also can help develop novel therapies for many neurological disorders exhibiting altered synaptic transmission, and is also relevant to diseases arising from defects in regulated secretion, including hypertension, cancer, and diabetes (27, 28).
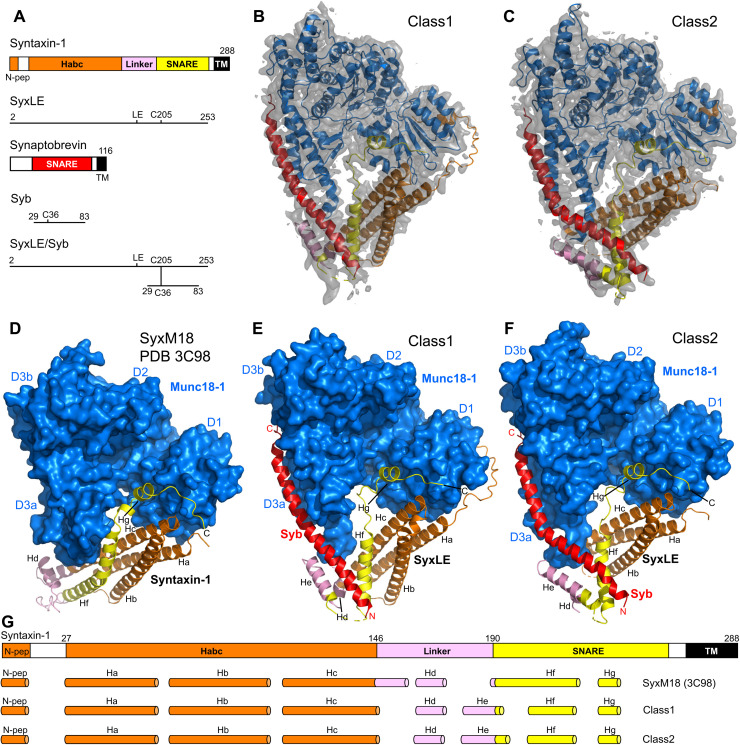
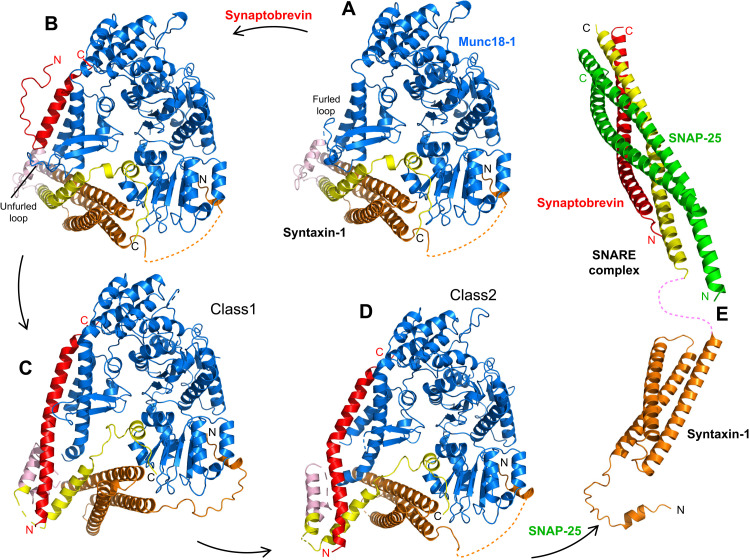
Fig. 1. Two cryo-EM structures of the template complex.
(A) Domain diagrams of syntaxin-1 and synaptobrevin, and summary of the fragments used to prepare SyxLE/Syb. N-pep, N-peptide; SNARE, SNARE motif. (B and C) 3D reconstructions of two structures of the template complex, class1 (A) and class2 (B), and corresponding ribbon diagrams fitted into the cryo-EM maps at 3.7 and 3.5 Å, respectively. (D to F) Comparison of the crystal structure of the syntaxin-1–Munc18-1 complex (SyxM18) (PDB code 3C98) (8, 34) (D) with the two cryo-EM structures of the template complex, class1 (E) and class2 (F). The surface of Munc18-1 is shown in blue, and the SNAREs are represented by ribbon diagrams with synaptobrevin (Syb) in red and syntaxin-1 in orange (N-peptide and Habc domain), pink (linker), and yellow (SNARE motif). The domains of Munc18-1 (D1, D2, D3a, and D3b) are labeled. The helices formed by syntaxin-1 (named Ha-Hg) are indicated. The N and C termini of Syb, as well as the C terminus of SyxLE, are labeled. (G) Summary of the locations of the helices observed in SyxM18, class1, and class2. The helices are represented by cylinders, and they are shown below a domain diagram of syntaxin-1 with selected residue numbers above to indicate domain boundaries. The helix formed by the N-peptide at the very N terminus is named N-pep, and subsequent helices of class1 and class2 are named Ha to Hg. To facilitate comparisons, the same nomenclature is used for SyxM18 helices in the same or similar positions as those observed in class1 and class2, but not that there is no He helix in SyxM18.
In addition to binding tightly to closed syntaxin-1, Munc18-1 interacts weakly with synaptobrevin (29), which suggested that Munc18-1 forms a template for SNARE complex assembly (24). This notion was firmly established by seminal crystal structures of the yeast vacuolar Sec1-Munc18 (SM) homolog Vps33 bound to the SNARE motif of the syntaxin-1 homolog Vam3 or the synaptobrevin homolog Nyv1 (30), which showed that simultaneous binding to Vps33 would place Vam3 and Nyv1 near each other in the correct register for SNARE complex assembly. Subsequent biophysical studies of Munc18-1 further supported the idea that a template complex of Munc18-1 bound to syntaxin-1 and synaptobrevin is a central intermediate between the Munc18-1–closed syntaxin-1 complex and the SNARE complex, and showed that the syntaxin-1 N-terminal region plays a crucial role in template complex formation (31, 32). However, the basis for such crucial role is unknown, and the steps that lead from the Munc18-1–closed syntaxin-1 complex to the SNARE complex remain enigmatic, in part because no structures are available for Munc18-1 bound to synaptobrevin or for any SM protein bound simultaneously to its two cognate SNAREs.
Here, we fill this fundamental gap, describing the elucidation of two structures of a synaptobrevin–Munc18-1–syntaxin-1 template complex by cryo–electron microscopy (cryo-EM). The two structures reveal how syntaxin-1 opens while Munc18-1 holds synaptobrevin and syntaxin-1 through the C termini of their SNARE motifs, favoring interactions between the SNARE motif N termini that are nucleated by the linker sequence between the syntaxin-1 SNARE motif and the Habc domain. Mutagenesis, together with SNARE complex assembly assays and reconstituted proteoliposome fusion experiments, supports the functional relevance of the cryo-EM structures. Our results explain the critical role of the syntaxin-1 N-terminal region and suggest a model whereby this region, together with Munc18-1, helps nucleate SNARE complex assembly via a mechanism involving multiple energy barriers that provide varied opportunities for tight regulation of neurotransmitter release.
RESULTS
Formation of a stable template complex
The weak affinity of synaptobrevin for Munc18-1 (29) hinders structural studies of complexes between them. To overcome this problem, we cross-linked the syntaxin-1 cytoplasmic region containing the LE mutation (SyxLE) to a synaptobrevin fragment spanning most of its SNARE motif (Syb) through a disulfilde bond between single cysteine residues placed at the N termini of their SNARE motifs (residue 36 of synaptobrevin and residue 205 of syntaxin-1). We refer to the resulting conjugate as SyxLE/Syb (Fig. 1A). Cross-linking through these residues was previously shown to support SNARE complex assembly (33) and to facilitate formation of a synaptobrevin–Munc18-1–syntaxin-1 template complex (32). To further favor template complex formation, we used Munc18-1 bearing a D326K gain-of-function mutation that increases the affinity of synaptobrevin for Munc18-1 because it unfurls a Munc18-1 loop that covers the synaptobrevin binding site (31). SyxLE/Syb coeluted with Munc18-1 D326K in gel filtration (fig. S1, A and B) and formed an SDS-resistant SNARE complex with SNAP-25 in the presence of Munc18-1 much more efficiently than the separate SyxLE and Syb fragments (compare the amounts of SNARE complex formed in lanes 6 and 9 in fig. S1C).
Cryo-EM structures of the template complex
Imaging of the template complex by cryo-EM and initial three-dimensional (3D) classification led to the identification of two major classes, which we refer to as class1 and class2 and were resolved at 3.7 and 3.5 Å, respectively (Fig. 1, B and C, figs. S2 and S3, and table S1). The two structures reveal extensive interactions of Munc18-1 with syntaxin-1 and synaptobrevin (see buried surface areas in table S2) and have similar architectures, with common features that were also observed in the crystal structure of the syntaxin-1–Munc18-1 complex (8) but with important differences (Fig. 1, D to F, and fig. S4).
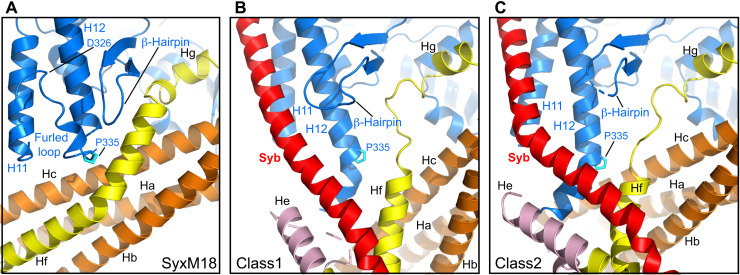
Munc18-1 has an almost identical three-domain arch shape in the three structures. The most notable difference is in the loop joining helices H11 and H12 of domain 3a, which is furled in the Munc18-1–syntaxin-1 complex, covering the synaptobrevin binding site, but is unfurled in class1 and class2 to allow synaptobrevin binding (Fig. 2). Almost the entire synaptobrevin SNARE motif forms a continuous helix in both class1 and class2, but in class2, the helix is bent at the middle (Fig. 1, E and F). The C-terminal half of the synaptobrevin SNARE motif binds to a groove formed by helices H11 and H12 of Munc18-1 (Figs. 1, E and F, and 2, B and C), in an analogous position to that observed for Nyv1 bound to Vps33 (fig. S5, A to C) (30). Binding to the groove is mediated by a combination of hydrophobic and polar interactions, and side chains in the interface include L307 and L348 of Munc18-1 (fig. S6, A to D), which had been implicated in synaptobrevin binding and/or template complex formation through mutagenesis (24, 32). The R56 side chain in the middle of the synaptobrevin SNARE motif binds to a pocket at the bottom of the H11-H12 groove in both class1 and class2, but there are almost no further contacts between Munc18-1 and residues N-terminal to R56 (fig. S6, E to H). This observation contrasts with the extensive contacts of the N-terminal half of Nyv1 with Vps33 in their complex (fig. S5A). Instead, the N-terminal half of the synaptobrevin SNARE motif interacts with syntaxin-1 (Fig. 1, E and F, and fig. S6, E to H), which is expected to be favored by the disulfide bond. It is unclear whether, during the process of SNARE complex assembly, the N-terminal half of the synaptobrevin SNARE motif might bind at some point to the unfurled loop of Munc18-1, as observed in the Vps33-Nyv1 complex (fig. S5A), but it seems most likely that such binding is transient if it occurs, as it prevents interactions between the N termini of the synaptobrevin and syntaxin-1 SNARE motifs for initiation of SNARE complex assembly. It is also worth noting that R56 of synaptobrevin forms a polar layer in the middle of the SNARE complex four-helix bundle, which otherwise is formed by hydrophobic layers (3), and is at the corner where the synaptobrevin helix bends in the class2 structure (fig. S6H). Thus, R56 may constitute a pivot point where helices formed by the N- and C-terminal halves of the synaptobrevin SNARE motif may change direction during SNARE assembly.
Fig. 2. Structural changes in Munc18-1 that lead to synaptobrevin binding and template complex formation.
(A to C) Close-up views of the area where the Munc18-1 loop unfurls to allow synaptobrevin binding in the syntaxin-1–Munc18-1 complex (SyxM18) before unfurling (A) and in class1 (B) and class2 (C), where the loop is unfurled and synaptobrevin is bound. Munc18-1 is colored in blue, synaptobrevin (Syb) in red, and syntaxin-1 in orange (N-peptide and Habc domain), pink (linker), and yellow (SNARE motif). P335 is shown as a stick model, with carbon atoms in cyan. The positions of the furled loop and D326 in the Munc18-1–syntaxin-1 complex, as well as of synaptobrevin (Syb), P335, a nearby β-hairpin, and selected helices of Munc18-1 and syntaxin-1 are indicated.
Some of the interactions of Munc18-1 with syntaxin-1 observed in class1 and class2 are analogous to those observed previously in the syntaxin-1–Munc18-1 complex, while other interactions are remodeled, resulting in slightly larger buried surface areas between Munc18-1 and syntaxin-1 in the two cryo-EM structures than in the binary syntaxin-1–Munc18-1 complex (table S2). This remodeling occurs at least in part because of large conformational changes required to initiate syntaxin-1 opening. Because of these conformational changes, the number of syntaxin-1 helices is different in the two cryo-EM structures compared to the Munc18-1–syntaxin-1 complex. To facilitate comparisons, we use the same nomenclature for the syntaxin-1 helices of class1, class2, and the Munc18-1–syntaxin-1 complex, but note that the latter is missing one helix (helix He; see Fig. 1G).
In both class1 and class2, syntaxin-1 forms a short α-helix at the very N terminus (the N-peptide motif) that binds to the N-terminal domain of Munc18-1, as in the syntaxin-1–Munc18-1 complex (34), and seven additional helices (called Ha-Hg) (Fig. 1, E and F, and fig. S4, B and C). The C terminus of the syntaxin-1 SNARE motif, which includes the short Hg helix, binds to a groove of domain 1 of Munc18-1 and a pocket that defines its arch shape, also as observed in the Munc18-1–syntaxin-1 complex (Fig. 1, D to F, and fig. S7, A to C). This interaction and that of the N-peptide are the most conserved among the multiple interactions between syntaxin-1 and Munc18-1 in their binary complex, class1 and class2, suggesting that they serve as anchor points that retain syntaxin-1 bound to Munc18-1, while other regions undergo substantial rearrangements in the pathway toward SNARE complex assembly. Note that the interaction of the C-terminal half of syntaxin-1 SNARE motif with the Munc18-1 pocket was also observed in the crystal structure of the vacuolar Vam3-Vps33 complex (30) (fig. S5D), suggesting that this interaction is universally conserved among SM proteins and their cognate SNARE from the syntaxin family.
The structure of the Habc domain of syntaxin-1 is very similar in class1, class2, and the syntaxin-1–Munc18-1 complex, and the Habc domain–Munc18-1 interface is similar in the three structures (fig. S7, D to F), but the orientation of Habc with respect to Munc18-1 is somewhat different in class1 (fig. S4, D to F). In contrast, comparisons with the crystal structure of the yeast SM protein Vps45 bound to the syntaxin-1 homolog Tlg2 (35), in which Tlg2 is open, show that the orientation of the Habc domain of Tlg2 resembles more that observed in class1 than that of class2 (fig. S5, G to I). These observations suggest that the Habc–Munc18-1 interface, and likely Habc-SM protein interfaces in general, can adapt to structural rearrangements that occur during formation of the template complex and subsequent events that lead to SNARE complex assembly.
The N-terminal half of the syntaxin-1 SNARE motif forms a long α-helix (Hf) that contacts domain 3a of Munc18-1 in both class1 and class2 (Fig. 1, E and F), and is in an analogous position to that of the homologous region of Vam3 in its complex with Vps33 (fig. S5, D to F). This region of syntaxin-1 also binds to the same site of domain 3a of Munc18-1 in the binary complex, but establishing more extensive contacts than in class1 and class2 (Fig. 2, A to C, and fig. S6, I to K). Moreover, the helix formed by this region is shifted and rotated in class1 and class2 with respect to its position in the Munc18-1–syntaxin-1 complex (fig. S6, I to K), where the helix is extended to the very N terminus of the SNARE motif, exhibiting a bend in the middle (Fig. 1D) that is not observed in helix Hf of class1 and class2 (Fig. 1, E and F). This feature of the Munc18-1–syntaxin-1 complex allows extensive interactions of the SNARE motif with the Habc domain that define the closed conformation of syntaxin-1. These contacts are less extensive in class1 and even less extensive in class2 (Fig. 3, A to C; fig. S7, H to J; and table S2), suggesting that class1 occurs first and class2 later in the pathway that leads from the Munc18-1–closed syntaxin-1 complex to the SNARE complex. Thus, the gradual rotation of the SNARE motif with respect to the Habc domain observed when comparing the three structures (Fig. 3, A to F, and fig. S7, H to J) may reflect how the syntaxin-1 conformation opens during this pathway.
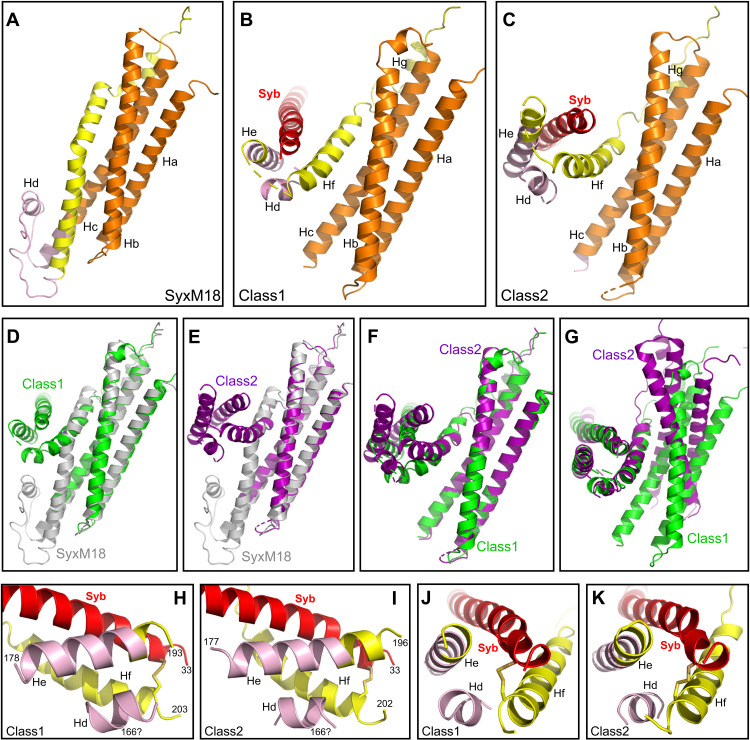
Fig. 3. Conformational changes in syntaxin-1 that lead to template complex formation.
(A to C) Close-up views of the region where the syntaxin-1 SNARE motif interacts with the Habc domain and the linker in SyxM18 (A) as well as with synaptobrevin in class1 (B) and class2 (C). (D to F) Superpositions among SyxM18 (gray), class1 (green), and class2 (purple) in the same close-up views shown in (A) to (C). The structures were superimposed using the atoms of the Habc domain. (G) Superposition of class1 and class2 in a similar close-up view but superimposing the atoms of the residues of the syntaxin-1 and synaptobrevin SNARE motifs (residues 206–219 and 37–50, respectively) that form the small four-helix bundle with helices Hd and He of the syntaxin-1 linker, showing that the small four-helix bundle has a similar structure in class1 and class2. Munc18-1 is not shown for simplicity in (A) to (G). (H to K) Close-up views of the small four-helix bundle in two different orientations. The views of (J) and (K) are rotated 90° with respect to those of (H) and (I). The N-terminal residues of the synaptobrevin SNARE motif and helix Hf formed by the SNARE motif of syntaxin-1, as well as the N- and C-terminal residues of helix He formed by the syntaxin-1 linker, are indicated. The sequence of the linker forming helix Hd could not be assigned, but we speculate that it corresponds to the same sequence that forms helix Hd in the Munc18-1–closed syntaxin-1 complex. The position of residue 166 resulting from this assumption is indicated. In (H) to (K), Munc18-1, the Habc domain, and the N-peptide are not shown for simplicity.
The local resolution of the density corresponding to the linker sequence connecting the Habc domain with the SNARE motif is worse than that of the rest of the complex for both class1 and class2 (see the bottom of the cryo-EM maps in the perspective shown in fig. S2B). Although we could not model the chains corresponding to part of the densities in this region, two helices were clearly distinguishable in both class1 and class2 (helices Hd and He; fig. S8). Helix He could be assigned to the sequence spanning the very N terminus of the syntaxin-1 SNARE motif and the preceding sequence from the linker in both structures, but the sequence corresponding to helix Hd could not be assigned and was modeled as polyalanine (Fig. 3, H and I, and fig. S8). On the basis of the distances from the C terminus of helix Hc to the N terminus of helix Hd and from the C terminus of helix Hd to the N terminus of helix He, we speculate that helix Hd of class1 and class2 is formed by the same residues of the linker that form helix Hd in the closed conformation of syntaxin-1 bound to Munc18-1 (Fig. 3A). Regardless of which specific residues form helix Hd, it is clear that the structure of the linker sequence is drastically different in class1 and class2 compared to the syntaxin-1–Munc18-1 complex (Figs. 1, D to F, and 3, A to F).
In both class1 and class2, helices Hd and He of the linker form a small four-helix bundle with the N terminus of the synaptobrevin SNARE motif and helix Hf, which comprises part of the syntaxin-1 SNARE motif N terminus (Fig. 3, H to K). This four-helix bundle is similar in class1 and class2 (Fig. 3G) and involves multiple contacts between side chains of the synaptobrevin and syntaxin-1 SNARE motifs (fig. S6, E to F) that are also in contact in the SNARE complex (3). It is noteworthy that formation of helix Hd requires considerable stretching of the residues connecting helix Hc to helix Hd and/or the residues connecting Hd to He (depending on which specific residues form Hd). Hence, it seems likely that this region is highly dynamic, and there is a range of structures in the particles used to construct the class1 and class2 cryo-EM maps such that helix Hd, helix He, and/or the C terminus of Hc (where the local resolution is also low) are stretched to different extents in some of the particles. Thus, formation of the small four-helix bundle may be transient, but it is remarkable that similar four-helix bundles are formed in class1 and class2 (Fig. 3, G to K) despite considerable differences in the structures of class1 and class2 around this region, including the bend of the synaptobrevin helix in class2 but not class1 (Figs. 1, E and F, and 2, B and C) and the different orientations of the short four-helix bundle with respect to the Habc domain (Fig. 3, B, C, and F). These observations suggest that this small four-helix bundle constitutes a relatively stable (or metastable) structure that helps to nucleate the first interactions between syntaxin-1 and synaptobrevin for eventual SNARE complex formation, while Munc18-1 provides a platform that holds the C-terminal halves of the syntaxin-1 and synaptobrevin SNARE motifs. This model explains why the syntaxin-1 N-terminal region is crucial for template complex formation (32).
Alteration of Munc18-1–SNARE interactions by mutagenesis
To study the functional effects of disrupting the template complex and hence test the functional relevance of our cryo-EM structures, we used a battery of Munc18-1 mutations that included the gain-of-function D326K and P335A mutations. The P335A mutation was previously designed to extend helix H12 and increase synaptobrevin binding, but such increase was not observed (24); instead, the P335A mutation decreased syntaxin-1 binding (26). Note that this mutation was also found to stabilize the synaptobrevin–Munc18-1–syntaxin-1 template complex (32). To impair binding of Munc18-1 to syntaxin-1 at a different site, we used an S42Q mutation expected to disrupt interactions with the syntaxin-1 SNARE motif C terminus. We also included the L307R and L348R mutations that disrupt synaptobrevin binding and/or template complex formation (24, 32), an E352K mutation that we designed to also disrupt Munc18-1–synaptobrevin binding, and a Q301D mutation that impairs neurotransmitter release and was reported to impair binding of Munc18-1 to synaptobrevin or to Munc13-1 (36, 37). The locations of the mutated residues are shown in Fig. 2A and figs. S6 (A to D) and S7 (A to C) (note that D326 is visible in the Munc18-1–syntaxin complex, but not in class1 and class2).
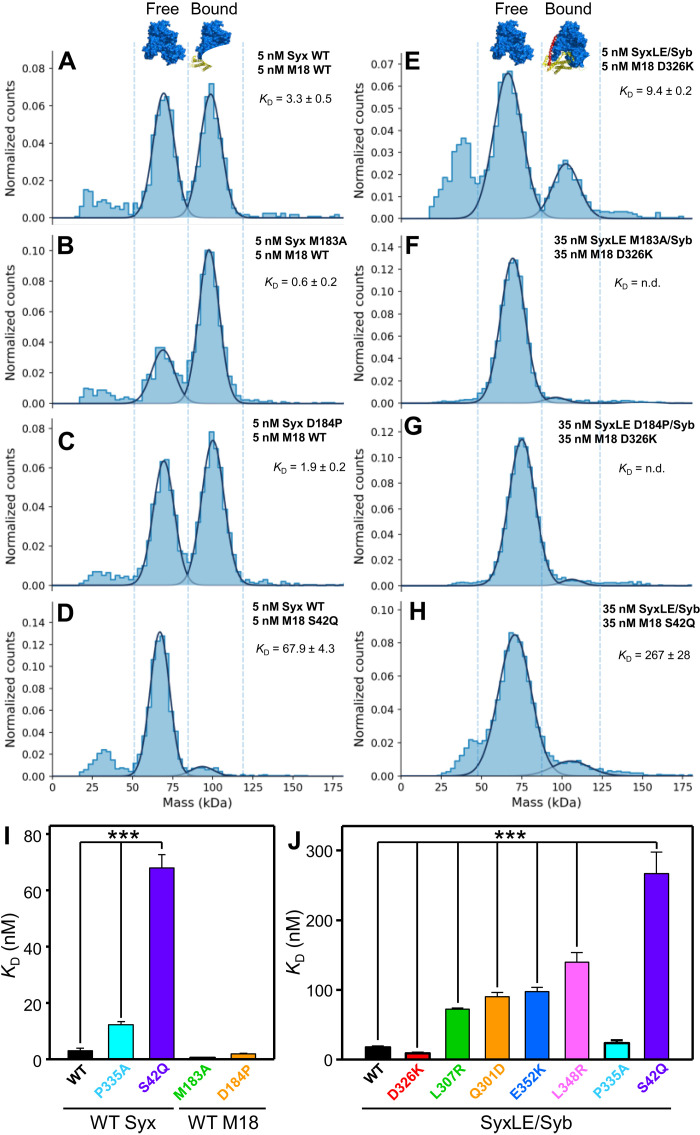
To analyze the effects of these mutations on binding of Munc18-1 to the syntaxin-1 cytoplasmic region and SyxLE/Syb, we used mass photometry, a single-molecule interferometric scattering–based technique that allows counting of proteins and complexes existing in solution (Fig. 4 and table S3) (38). This method yields reliable measurements of molecular masses in the 30-kDa to 5-MDa range and hence allowed us to count the numbers of free Munc18-1 molecules and Munc18-1–syntaxin-1 or Munc18-1–SyxLE/Syb complexes in samples containing mixtures of Munc18-1 with syntaxin-1 or SyxLE/Syb, respectively. The dissociation constants (KD’s) could thus be calculated from these counts. We measured a KD of 3.3 nM between Munc18-1 and syntaxin-1, consistent with previous isothermal titration calorimetry data (34), whereas the KD of Munc18-1 for SyxLE/Syb was 17.9 nM. It is worth noting that the syntaxin-1 LE mutant by itself binds to Munc18-1 with low nanomolar affinity (34), which can be attributed to the fact that, although the LE mutation destabilizes the closed conformation (7), this syntaxin-1 mutant can still adopt a closed conformation upon binding to Munc18-1 (39). This conformation is stabilized by the L205 side chain, which is buried and interacts with the Habc domain, but these interactions are prevented by the L205C mutation and the cross-link to synaptobrevin in SyxLE/Syb, which thus cannot adopt a closed conformation. Hence, it is not surprising that SyxLE/Syb binds to Munc18-1 with somewhat weaker affinity than the Syx LE mutant.
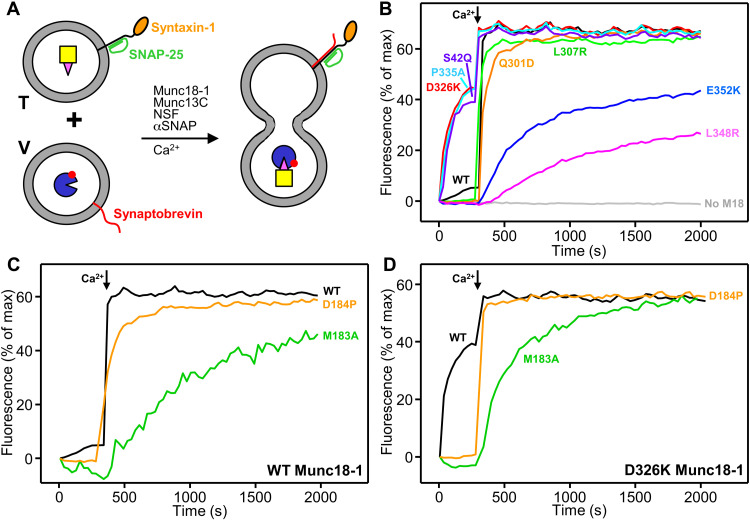
Fig. 4. Analysis of Munc18-1–SNARE interactions by mass photometry.
(A to H) Normalized histograms of mass distributions observed for samples containing the indicated concentrations of WT or mutant Munc18-1 (M18) plus WT or mutant syntaxin-1 (2–253) (Syx) (A to D), or mutant Munc18-1 plus SyxLE/Syb with or without the M183A or D184P mutations (E to H). Gaussian fits (solid lines) were used to calculate the populations of free and bound Munc18-1 and derive dissociation constants (KD’s). Binding of Munc18-1 D326K to SyxLE M183A/Syb and SyxLE D184P/Syb was too weak to derive reliable KD’s. (I and J) Bar diagrams illustrating the average KD’s (table S3) obtained from six independent experiments for samples containing WT or mutant Munc18-1 plus WT or mutant syntaxin-1 (2–253) (I), or WT or mutant Munc18-1 plus SyxLE/Syb with or without the M183A or D184P mutations (J). Error bars represent SDs. Statistical significance and P values were determined by one-way analysis of variance (ANOVA) with the Holm-Sidak test (***P < 0.001).
The Munc18-1 P335A mutation induced Munc18-1 dimerization, but we were able to distinguish the peak of the syntaxin-1–Munc18-1 P335A heterodimer (fig. S9) and obtained a KD of 12.3 nM, confirming that this mutation impairs syntaxin-1 binding (26). However, P335A had no overt effect on Munc18-1 binding to SyxLE/Syb (Fig. 4J). These results are consistent with the fact that P335 packs closely against syntaxin-1 in the Munc18-1–syntaxin-1 complex, whereas P335 does not contact syntaxin-1 in class1 and class2 (fig. S6, I to K). In contrast, the S42Q mutation strongly impaired binding of Munc18-1 to both syntaxin-1 and SyxLE/Syb (Fig. 4, D and H to J), which indicates that the C-terminal half of the syntaxin-1 SNARE motif plays a key role in binding to Munc18-1 both in the binary complex and in the template complex, and is consistent with the similarity of the binding modes observed in the corresponding region of the binary complex, class1 and class2 (fig. S7, A to C). Binding of Munc18-1 to SyxLE/Syb was strengthened by the D326K mutation, as expected, and was impaired to different degrees by the L307R, Q301D, E352K, and L348R mutations (Fig. 4J). These results are consistent with our cryo-EM structures and support the notion that the physiological effects of the Q301D mutation arise from impairment of Munc18-1 binding to synaptobrevin (37) rather than to Munc13-1 (36).
Disrupting the template complex impairs trans-SNARE complex formation and fusion
To test the effects of the Munc18-1 mutations on SNARE complex assembly, we first adapted a solution fluorescence resonance energy transfer (FRET) assay that showed that the Munc13-1 MUN domain accelerates the transition from the Munc18-1–syntaxin-1 complex to the SNARE complex (11). To avoid using the high concentrations of MUN domain required for such acceleration, we covalently attached the MUN domain to the C terminus of the syntaxin-1 cytoplasmic region with a long flexible linker (Fig. 5A). The syntaxin-1–MUN domain fusion (SyxMUN) assembled into SNARE complexes with SNAP-25 and soluble synaptobrevin as efficiently as syntaxin-1 alone (Fig. 5B). Binding of Munc18-1 to syntaxin-1 alone abolished SNARE complex assembly, but the SyxMUN fusion bound to Munc18-1 was still able to assemble into SNARE complexes (Fig. 5B). The reaction was stimulated by the P335A and S42Q Munc18-1 mutations, but not by D326K, and slight accelerations also appeared to be caused by the mutations that disrupt synaptobrevin binding (Fig. 5C, and fig. S10A). The P335A and S42Q mutations allowed SNARE complex assembly even in the absence of the MUN domain (Fig. 5D and fig. S10B). These results are consistent with an involvement of the template complex in SNARE complex assembly, as the rates of assembly are affected by mutations that disrupt interactions of Munc18-1 with syntaxin-1 and synaptobrevin. However, the observation that these mutations accelerate assembly while D326K does not have an effect shows that the rate-limiting step for SNARE complex assembly in these solution assays is not the binding of synaptobrevin to the Munc18-1–syntaxin-1 complex to form the template complex but rather the dissociation of the syntaxin-1 and synaptobrevin SNARE motifs from Munc18-1.
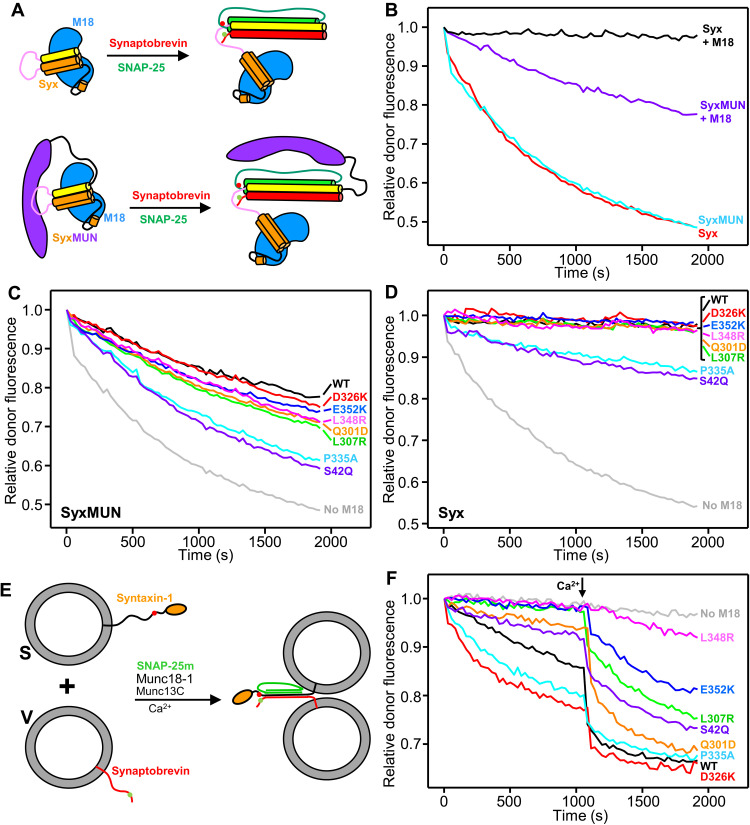
Fig. 5. Effects of mutations in Munc18-1 on SNARE complex assembly rates.
(A) Diagram summarizing the assays used to monitor SNARE complex assembly in solution starting with Munc18-1 (blue) bound to the syntaxin-1 cytoplasmic region (Habc domain, orange; linker, pink; SNARE motif, yellow) alone (Syx) or covalently linked to the Munc13-1 MUN domain (purple) (SyxMUN). Synaptobrevin (red) was labeled with a donor fluorescent probe (green dot), and SNAP-25 (green) was labeled with an acceptor probe (red dot). SNARE complex assembly was monitored by the development of FRET between the probes. (B to D) Decrease in relative donor fluorescence (normalized to the first time point) as a function of time in SNARE complex assembly reactions where synaptobrevin and SNAP-25 were added to Syx or SyxMUN free or pre-bound to WT Munc18-1 (M18) (B), to SyxMUN bound to WT or mutant Munc18-1 (C), or to Syx bound to WT or mutant Munc18-1 (as indicated) (D). (E) Diagram summarizing trans-SNARE complex assembly assays between V-liposomes containing synaptobrevin labeled with a donor fluorescent probe (green dot) and S-liposomes containing syntaxin-1 labeled with an acceptor probe (red dot) in the presence of Munc13C, a SNAP-25 mutant (SNAP-25m), and WT or mutant Munc18-1. (F) Decrease in relative donor fluorescence (normalized to the first time point) as a function of time in trans-SNARE complex assembly assays performed with WT or mutant Munc18-1 as indicated. Reactions were initiated in the presence of 100 μM EGTA, and Ca2+ (600 μM) was added at the time indicated by the arrow.
To better mimic the geometry of SNARE complex assembly in neurons, we next used a FRET assay that measures assembly of trans-SNARE complexes between synaptobrevin-containing liposomes (V-liposomes) and syntaxin-1 liposomes (S-liposomes) (Fig. 5E), adapting a previously described assay that used SNAP-25 with an M71D-L78D mutation in the C terminus of its first SNARE motif (SNAP-25m) to prevent liposome fusion (10). The S-liposomes were incubated with wild-type (WT) or mutant Munc18-1, and trans-SNARE complex assembly was monitored upon addition of V-liposomes, SNAP-25m, and a fragment spanning the conserved C-terminal region of Munc13-1, which includes its C1, C2B, MUN, and C2C domains (Munc13C). As expected (10), assembly in the presence of WT Munc18-1 was slow in the absence of Ca2+ and was strongly activated by Ca2+, which likely arises because Ca2+ binding to the Munc13–1 C2B domain changes the orientation in which Munc13C bridges the two membranes (40). The D326K and P335A mutations substantially enhanced Ca2+-independent trans-SNARE complex assembly (Fig. 5F and fig. S10C), consistent with the gains of function observed previously for these two mutants (16, 31). However, the S42Q mutation impaired trans-SNARE complex assembly, in contrast to the effect observed for this mutation in the solution assays. This opposite effects might arise because of differences in the transition states that determine the reaction rates in the two assays, but further research will be required to test this proposal. The Q301D, L307R, L348R, and E352K mutations decreased assembly in the absence and presence of Ca2+ to different extents (Fig. 5F and fig. S10, C and D) that approximately correlated with impairment in binding of Munc18-1 to SyxLE/Syb (Fig. 4J).
To analyze how the mutations affect the ability of Munc18-1 to support SNARE-dependent membrane fusion, we used an assay that measures content mixing between V-liposomes and liposomes containing syntaxin-1 and SNAP-25 (T-liposomes) in the presence of Munc18-1, Munc13C, NSF, and αSNAP (13) and that has allowed us to establish many correlations between the effects of mutations on liposome fusion and their effects on neurotransmitter release in neurons (Fig. 6A) (13, 14, 40, 41). As expected, no liposome fusion was observed in the absence of Munc18-1, and inclusion of WT Munc18-1 yielded inefficient Ca2+-independent fusion that was strongly stimulated by addition of Ca2+ (Fig. 6B). The D326K and P335A mutations strongly enhanced Ca2+-independent fusion, as observed previously (16, 31), and the S42Q mutation induced a similar stimulation (Fig. 6B and fig. S11A). In contrast, the Q301D, L307R, L348R, and E352K mutations abolished the small amount of Ca2+-independent fusion observed for WT Munc18-1 and impaired Ca2+-dependent fusion to different extents (Fig. 6B and fig. S11, A and B) that approximately correlate with the effects of these mutations on binding of Munc18-1 to SyxLE/Syb and on trans-SNARE complex assembly (Figs. 4J and 5F and fig. S10, C and D). Although the effects of the L307R and Q301D mutations on Ca2+-dependent fusion appear to be small, they are significant, as mild impairments in activity in these assays are often not observable (40).
Fig. 6. Effects of Munc18-1 and syntaxin-1 mutations on liposome fusion.
(A) Diagram summarizing the content mixing between T-liposomes containing syntaxin-1 and SNAP-25, which were preincubated with Munc18-1, NSF, and αSNAP, and V-liposomes containing synaptobrevin in the presence of Munc13C. V-liposomes contain trapped Cy5-strepatavidin, and T-liposomes contain trapped PhycoE-biotin. (B to D) Content mixing between V-liposomes and T-liposomes was monitored from the increase in the fluorescence signal of Cy5-streptavidin caused by FRET with PhycoE-biotin. Assays were performed with WT or mutant Munc18-1 (B), or WT or mutant syntaxin-1 and WT (C) or D326K mutant (D) Munc18-1, as indicated. Experiments were started in the presence of 100 μM EGTA, and Ca2+ (600 μM) was added at 300 s.
The formation of a small four-helix bundle by synaptobrevin, the syntaxin-1 SNARE motif, and helices Hd and He from the syntaxin-1 linker in class1 and class2 (Figs. 1, E and F, and 3, H to K) suggests that the linker acts as a template to nucleate assembly of the SNARE complex. To test the functional relevance of the structure formed in this region, we designed two mutations in helix He: a D184P mutation expected to disrupt the helical structure and an M183A mutation designed to disrupt hydrophobic contacts that stabilize the short four-helix bundle (fig. S6, E to H) and involve a highly conserved residue (fig. S12). These mutations are not expected to substantially perturb the closed conformation of syntaxin-1 because these residues are exposed in a loop (fig. S7G). The M183A and D184P mutations did not impair syntaxin-1 binding to Munc18-1, but they strongly disrupted binding of SyxLE/Syb to Munc18-1 D326K (Fig. 4, A to C and E to G).
Ca2+-dependent liposome fusion in the presence of WT Munc18-1 was strongly disrupted by the M183A mutation and was impaired by the D184P mutation (Fig. 6C and fig. S11C). Analysis of the liposomes by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) (fig. S11E) showed that these results did not arise from poor syntaxin-1 incorporation onto the liposomes. Since the high efficiency of Ca2+-dependent fusion might have masked the severity of the effect of the D184P mutation, we also performed liposome fusion assays in the presence of the Munc18-1 D326K mutant, which yields Ca2+-independent fusion. Both M183A and D184P abolished Ca2+-independent fusion (Fig. 6D and fig. S11D), emphasizing the importance of the He helix of syntaxin-1 for the molecular events that lead to liposome fusion.
DISCUSSION
The crystals structures of Vps33-SNARE complexes (30) and multiple data on Munc18-1 (24, 31, 32) showed that SM proteins form templates for SNARE complex assembly and that a synaptobrevin–Munc18-1–syntaxin-1 template complex represents a crucial intermediate in the path from the Munc18-1–closed syntaxin-1 complex to the SNARE complex. However, the steps leading to SNARE complex assembly and the basis for the key role played by the syntaxin-1 N-terminal region in template complex formation remained poorly understood, and no structure was available for Munc18-1 bound to synaptobrevin or for any SM protein bound to its two cognate SNAREs. Our cryo-EM structures now fill this gap and, together with our biophysical assays and previous data, allow us to develop a realistic model of the steps that lead from the Munc18-1–closed syntaxin-1 complex to the SNARE complex in which the syntaxin-1 linker plays a key role in nucleating SNARE assembly, while the C termini of the syntaxin-1 and synaptobrevin SNARE motifs are held at distant sites of Munc18-1.
Structural characterization of the template complex was hindered by its necessarily dynamic nature as an intermediate in the pathway to SNARE complex assembly. To overcome this problem, we used chemical cross-linking and took advantage of the ability of cryo-EM to characterize substantially populated states within conformational ensembles. In principle, the introduction of a disulfide bond between synaptobrevin and syntaxin-1 might cause local structural distortions, but multiple arguments suggest that the bias introduced by the cross-link selects for configurations of the ensemble that are on the productive pathway for SNARE complex assembly. First, the two cross-linked residues are in contact in the structure of the SNARE complex (3). Second, the cross-link facilitates SNARE complex assembly (fig. S1C) (33) and formation of the template complex in optical tweezer experiments (32). Third, these experiments established multiple correlations between the effects of mutations on template complex formation and their effects on liposome fusion or neurotransmitter release (32). Fourth, our cryo-EM structures readily explain our Munc18-1–SNARE binding, SNARE complex assembly, and liposome fusion data. Thus, the effects of Munc18-1 mutations on binding (Fig. 4) support the interaction modes of Munc18-1 with synaptobrevin and syntaxin-1 observed in the two structures. Moreover, the effects of the Munc18-1 mutations on SNARE complex assembly and fusion (Figs. 5 and 6) illustrate the importance not only of Munc18-1 binding to the syntaxin-1 and synaptobrevin SNARE motifs but also of release of these interactions to allow SNARE complex formation. Compelling evidence for the functional relevance of the conformations observed for the syntaxin-1 linker in our cryo-EM structures is provided by the severe disruption of Munc18-1–SyxLE/Syb binding and of liposome fusion caused by the M183A and D184P mutations in syntaxin-1 (Figs. 4 and 6, C and D).
Our results suggest a model for the transition from the Munc18-1–syntaxin-1 complex to the SNARE complex where states resembling our two cryo-EM structures of the template complex are at center stage (Fig. 7). Multiple energy barriers need to be overcome during this transition, for instance, to open the syntaxin-1 conformation, to unfurl the Munc18-1 loop, and to disrupt extensive Munc18-1–SNARE interactions that are expected to confer specificity to SNARE complex formation but need to be released for the SNARE complex to form fully. Hence, the transition must involve a series of steps. Interactions of the syntaxin-1 N-peptide and the Habc domain with Munc18-1 (Fig. 1, D to F, and fig. S4, A to C) are likely present throughout the pathway and may remain even after the SNARE complex is formed. These interactions, together with binding of the C-terminal half of the syntaxin-1 SNARE motif to the Munc18-1 cavity (Fig. 1, D to F, and fig. S4, A to C), provide anchor points to keep syntaxin-1 bound to Munc18-1, while the N-terminal half of the SNARE motif and the linker undergo conformational rearrangements that are necessary to form the template complex. At a distal site, binding of the C-terminal half of the synaptobrevin SNARE motif to the groove between helices H11 and H12 of Munc18-1 (Fig. 2, A to C) provides an anchor point for synaptobrevin. Thus, a central aspect of this model is that the attachment of the C-terminal halves of the synaptobrevin and syntaxin-1 SNARE motifs to distal parts of Munc18-1 allows their N-terminal halves to come close and bind, as proposed from optical tweezer experiments (32).
Fig. 7. Model of SNARE complex assembly templated by Munc18-1 and the syntaxin-1 N-terminal region.
The model postulates that assembly starts with Munc18-1 bound to closed syntaxin-1 (A) (PDB code 3C98) and is initiated when the Munc18-1 loop unfurls to allow synaptobrevin binding (B) (model built manually). (C and D) Conformational changes in syntaxin-1 that are stimulated by Munc13-1 (not shown) lead to template complex configurations such as those of class1 and class2, where syntaxin-1 gradually opens and the syntaxin-1 linker nucleates interactions between the syntaxin-1 and synaptobrevin SNARE motifs. The color code is the same as in Fig. 1. (E) Binding to SNAP-25 (green) eventually leads to formation of the SNARE complex (PDB codes: Habc domain 1BR0; SNARE four-helix bundle 1SFC). The N and C termini of the SNAREs are indicated in the relevant panels.
Unfurling the Munc18-1 loop to allow synaptobrevin binding (Fig. 2, A to C) is clearly a key event for SNARE complex assembly, as shown by the observation that the D326K mutation that unfurls the loop leads to gains of function in vivo (31) and in the trans-SNARE complex assembly and liposome fusion assays in vitro (Figs. 5F and 6B). However, this mutation or mutations that disrupt Munc18-1–synaptobrevin interactions did not affect SNARE complex assembly in our solution assays (Fig. 5C). Thus, synaptobrevin binding is a rate-limiting step in the trans-SNARE complex assembly, but not in the solution assays, which in principle is unexpected because bridging of the two membranes by Munc13-1C (14) should increase the local concentration of synaptobrevin and the Munc18-1–syntaxin-1 complex in the former assays. These findings likely arise because the highly elongated Munc13-1C fragment has a tendency to bridge the membranes in an approximately perpendicular orientation that keeps the membranes apart in the absence of Ca2+ (14, 40), which may hinder binding of synaptobrevin to the Munc18-1–syntaixn-1 complex. The effects of the D326K mutation are similar to those caused by the P335A mutation in Munc18-1, which also induces a gain of function in vivo (16, 25) and enhances trans-SNARE complex assembly and liposome fusion (Figs. 5F and 6B). The region around P335 and a nearby β-hairpin are well packed against syntaxin-1 in the Munc18-1–syntaxin-1 complex, but not in class1 and class2 (Fig. 2 and fig. S6, I to K). Correspondingly, the P335A mutation markedly impairs binding of Munc18-1 to syntaxin-1, but not to SyxLE/Syb (Fig. 4 and fig. S9). Thus, the gain of function caused by this mutation likely arises because it helps to release the interaction of the N-terminal half of the syntaxin-1 SNARE motif with Munc18-1, which is required to form the template complex and might occur in a concerted fashion with the unfurling of the Munc18-1 loop. However, the observation that P335A accelerates SNARE complex assembly in the solution assay (Fig. 5, C and D), unlike D326K, suggests that unfurling of the Munc18-1 loop occurs first (Fig. 7B), before release of the interactions between the region of Munc18-1 around P335 and syntaxin-1.
Interactions of the N-terminal half of the syntaxin-1 SNARE motif with the Habc domain, which define the closed conformation, also need to be released to allow binding between the N-terminal halves of the synaptobrevin and syntaxin-1 SNARE motifs to form the template complex (Fig. 3, A to C). Since the syntaxin-1 linker is packed against the SNARE motif in the closed conformation (Fig. 3A and fig. S7G), the motion of the SNARE motif to bind to synaptobrevin must also be accompanied by structural changes in the syntaxin-1 linker. Hence, multiple energy barriers need to be overcome for the synaptobrevin and syntaxin-1 SNARE motifs to come together, and it seems unlikely that the resulting interactions between a few residues of the SNAREs are sufficient to overcome these barriers. Even if such interactions occur transiently, the system might quickly revert to the closed syntaxin-1–Munc18-1 complex structure unless the initial synaptobrevin and syntaxin-1 interactions are stabilized. The SNARE motifs of syntaxin-1 and synaptobrevin form a short four-helix bundle with helices Hd and He from the syntaxin-1 linker in both class1 and class2 (Figs. 1, E and F, and 3, H to K), and the four-helix bundle includes native contacts between residues of synaptobrevin and syntaxin-1 that also interact in the SNARE complex (e.g., V42, M46 of synaptobrevin and L212, F216 of syntaxin-1; fig. S6, E and F). These observations suggest that formation of this small four-helix bundle stabilizes the interactions between the SNARE motifs and constitutes the initiation point for SNARE complex formation.
The different extents of interaction between the syntaxin-1 SNARE motif and the Habc domain observed in the Munc18-1–syntaxin-1 complex, class1, and class2 (Figs. 3, A to C, and 7, A, C, and D, and fig. S7, H to J) help to visualize how syntaxin-1 opens gradually and suggest that, once the small four-helix bundle forms, it helps to maintain the native synaptobrevin–syntaxin-1 contacts during the opening process. However, this structure is likely metastable, as discussed above, and helices Hd-He need to be at least partially dissociated in subsequent events to allow SNAP-25 binding to syntaxin-1 and synaptobrevin. As the SNARE complex zippers, the C-terminal halves of the synaptobrevin and syntaxin-1 SNARE motifs need to dissociate from Munc18-1 to allow full SNARE complex assembly (Fig. 7E). The central role of the syntaxin-1 linker in nucleating SNARE complex assembly postulated by this model is supported by the severe impairment in liposome fusion caused by the M183E and D184P mutations in the linker (Fig. 6, C and D). An 18-residue sequence upstream of the SNARE motif in the linker region of Vam3 was found to play a key role in SNARE complex assembly and liposome fusion mediated by the yeast vacuolar SNAREs (42), suggesting that linker sequence upstream of the SNARE motifs from the syntaxin family may play a universal role in nucleating SNARE complex formation. Note, however, that the homotypic fusion and protein sorting (HOPS) complex, which mediates yeast vacuolar fusion and contains Vps33, associates first with the synaptobrevin homolog Nyv1 and later with the syntaxin-1 homolog Vam3 (43, 44), in contrast with the initial binding of Munc18-1 to syntaxin-1 and later to synaptobrevin. Moreover, Vam3 does not adopt a closed conformation (45). These findings illustrate that there are also substantial differences in the molecular mechanisms underlying yeast vacuolar fusion and synaptic vesicle exocytosis.
The multiple energy barriers that hinder the transition from the Munc18-1–syntaxin-1 complex to the template complex and then to the SNARE complex provide varied possibilities for regulation. Munc13-1 acts as a master regulator of release (5) and is expected to influence the molecular events outlined above by at least two mechanisms. First, as mentioned above, the orientation of Munc13-1 as it bridges the two membranes controls the accessibility of the Munc18-1–syntaxin-1 complex to synaptobrevin, and changes in this orientation induced by Ca2+ and diacylglycerol during repetitive stimulation likely increase this accessibility, facilitating SNARE complex assembly (40). Second, Munc13-1 helps to open syntaxin-1 (11, 12) through weak interactions with the residues of the syntaxin-1 linker that connect helices Hc and Hd (46, 47). These interactions might facilitate formation of the small four-helix bundle involving the linker and the syntaxin-1 and synaptobrevin SNARE motifs, or may simply destabilize the linker conformation existing in the Munc18-1–syntaxin-1 complex, thus catalyzing conformational rearrangements in syntaxin-1. Both effects and perhaps direct Munc18-1–Munc13-1 interactions (36) might also stimulate unfurling of the Munc18-1 loop to facilitate synaptobrevin binding. Release is also modulated by kinases that phosphorylate Munc18-1, altering interactions with SNAREs [e.g., (48)]. Hence, this pathway is central for the exquisite regulation of synaptic vesicle fusion, which is a hallmark that allows presynaptic terminals to act as small computational units in the brain (49). The facts that a mutation in Unc-18 analogous to P335A partially rescues the severe phenotypes caused by deletion of Unc-13 (16) and that the LE mutation of syntaxin-1 can partially rescue the phenotypes caused by the absence of very diverse proteins (15, 22, 23) emphasize how understanding the mechanism of neurotransmitter release allows manipulation of the release efficiency. The structures of Munc18-1–SNARE complexes that are now available, together with the mechanistic understanding yielded by our mutagenesis studies (Figs. 4 to 6) and previous data [reviewed in (5)], provide a framework to design strategies for modulation of neurotransmitter release and other forms of regulated secretion that might have therapeutic use for a wide variety of diseases.
MATERIALS AND METHODS
Protein expression and purification
Escherichia coli expression and purification of full-length rat syntaxin-1A, the cytoplasmic fragment of rat syntaxin-1A (residues 2–253), a cysteine-free variant of full-length rat SNAP-25a, a full-length rat synaptobrevin-2, rat synaptobrevin-2 (residues 29–83), rat synaptobrevin (residues 1–96), full-length rat Munc18-1, full-length Cricetulus griseus NSF, full-length Bos taurus αSNAP, and a rat Munc13-1 fragment spanning the C1, C2B, MUN, and C2C domains (residues 529–1725, Δ1408–1452) (referred to as Munc13C) were described previously (9, 10, 13, 50–52). The following mutants were also described previously (10, 31, 47) and were purified through the same protocols used for the WT proteins: full-length syntaxin-1 S186C, C145A, C271A, C272A; SNAP-25a R136C C84S, C85S, C90S, C92S; SNAP-25a M71D, L78D C84S, C85S, C90S, C92S (SNAP-25m); full-length synaptobrevin-2 L26C; synaptobrevin-1 1–96 L26C; Munc18-1 P335A; Munc18-1 D326K; and Munc18-1 L348R.
Briefly, full-length syntaxin-1 was expressed overnight at 25°C upon induction with 0.4 mM isopropyl-β-d-thiogalactoside (IPTG). Cell pellets were resuspended with 20 mM Hepes (pH 7.4), 500 mM NaCl, 8 mM imidazole, and 1 mM tris(2-carboxyethyl)phosphine (TCEP). The protein was initially purified via affinity chromatography using HisPur Ni–nitrilotriacetic acid (NTA) resin (Thermo Fisher Scientific) in 20 mM tris (pH 7.4), 500 mM NaCl, 8 mM imidazole, 2% (v/v) Triton X-100, and 6 M urea. Upon extensive washes, the protein was eluted in 20 mM tris (pH 7.4), 500 mM NaCl, 400 mM imidazole, and 0.1% dodecylphosphocholine (DPC). The polyhistidine tag was removed using thrombin protease, followed by size exclusion chromatography on a Superdex 200 column (GE 10/300) equilibrated in 20 mM tris (pH 7.4), 125 mM NaCl, 1 mM TCEP, and 0.2% DPC.
Expression of syntaxin-1 2–253 was induced with 0.4 mM IPTG and expressed overnight at 25°C. Upon cell lysis, purification was done using Glutathione Sepharose 4B resin (GE) in phosphate-buffered saline (PBS; pH 7.4), PBS with 1% (v/v) Triton X-100, and PBS with 1 M NaCl. The glutathione S-transferase (GST)–tag was cleaved by thrombin protease, and the eluted protein was further purified by anion exchange chromatography on a HiTrap Q column (GE) in 25 mM tris (pH 7.4) and 1 mM TCEP using a linear gradient from 0 to 1 M NaCl. Please note that the purification of syntaxin-1 2–253 C145A, L165A, E166A, L205C (SyxLE) was carried out with the same procedure except that the final purification was done by size exclusion chromatography using a Superdex 75 (GE 10/300) column equilibrated with 100 mM phosphate buffer at pH 7.0 with 100 mM NaCl, 6 M urea, 0.2 mM EDTA, and 0.5 mM TCEP.
Cysteine-free SNAP-25 was expressed overnight at 25°C upon induction with 0.4 mM IPTG. Upon cell lysis, protein purification was performed using HisPur Ni-NTA resin (Thermo Fisher Scientific) in 50 mM tris (pH 8.0), 500 mM NaCl, 20 mM imidazole, and 1% (v/v) Triton X-100. The His6-tag was cleaved by thrombin protease, and the protein was purified by size exclusion chromatography using a Superdex 75 column (GE 16/60) in 50 mM tris (pH 8.0) and 150 mM NaCl.
Full-length synaptobrevin-2 was expressed overnight at 25°C upon induction with 0.4 mM IPTG. Cells were resuspended in PBS buffer containing 1% (v/v) Triton X-100. Purification was done using Glutathione Sepharose 4B resin (GE) at 4°C. The bound proteins were treated with PBS with 1% (v/v) Triton X-100, followed by the addition of thrombin to cleave the GST-tag. The protein was further purified by cation exchange chromatography on a HiTrap S column (GE) in 25 mM NaAc (pH 5.5), 1 mM TCEP, and 1% (w/v) using a linear gradient from 0 to 1 M NaCl.
Expression of synaptobrevin-2 1–96 or synaptobrevin-2 29–83 Q36C (Syb) was induced with 0.4 mM IPTG and expressed overnight at 23°C. Purification was done using Glutathione Sepharose 4B resin (GE), followed by cleavage of the GST-tag. The final purification of synaptobrevin-2 1–96 was performed using size exclusion chromatography on a Superdex 75 column (GE 16/60) equilibrated in 20 mM tris (pH 7.4) and 125 mM NaCl. The final purification of synaptobrevin-2 29–83 Q36C was carried out using size exclusion chromatography on a Superdex 75 column (GE 10/300) equilibrated with 100 mM phosphate buffer at pH 7.0 with 100 mM NaCl, 6 M urea, 0.2 mM EDTA, and 0.5 mM TCEP.
Expression of αSNAP was induced by the addition of 0.4 mM ITPG and continued overnight at 25°C. Protein purification was performed using Glutathione Sepharose 4B resin (GE) by washing bound proteins with PBS, PBS with 1% (v/v) Triton X-100, and PBS with 1 M NaCl. Upon GST-tag cleavage in the presence of thrombin, the protein was purified by size exclusion chromatography using a Superdex 75 column (GE 16/60) in 20 mM tris (pH 7.4), 150 mM KCl, and 1 mM TCEP.
Expression of NSF was induced with 0.4 mM IPTG and continued overnight at 20°C. Purification was performed using HisPur Ni-NTA resin (Thermo Fisher Scientific), followed by size exclusion chromatography of hexameric NSF on a Superdex S200 column (GE 16/60) in 50 mM tris (pH 8.0), 100 mM NaCl, 1 mM adenosine triphosphate (ATP), 1 mM EDTA, 1 mM dithiothreitol (DTT), and 10% (v/v) glycerol. Removal of the His6-tag and monomerization of NSF were performed using Tobacco Etch Virus (TEV) protease and apyrase, respectively, while dialyzing with nucleotide-free buffer for 36 hours. To separate the hexameric form of NSF from the monomeric, three rounds of size exclusion chromatography on a Superdex S200 column (GE 16/60) in 50 mM NaPi (pH 8.0), 100 mM NaCl, and 0.5 mM TCEP were performed by reinjecting fractions with hexameric NSF. Final reassembly of monomers and gel filtration chromatography of reassembled hexameric NSF were done using a Superdex S200 column (GE 16/60) in 50 mM tris (pH 8.0), 100 mM NaCl, 1 mM ATP, 1 mM EDTA, 1 mM TCEP, and 10% (v/v) glycerol.
Expression of full-length Munc18-1 was induced with 0.4 mM IPTG and continued overnight at 20°C. Upon cell lysis and centrifugation, the supernatant was loaded on Glutathione Sepharose 4B resin (GE) at 4°C, and the bound proteins were washed with PBS, PBS with 1% (v/v) Triton X-100, and PBS with 1 M NaCl. The GST-tag was cleaved by a thrombin treatment, followed by immediate size exclusion chromatography using a Superdex 200 column (GE 16/60) in a buffer containing 20 mM tris (pH 7.4), 200 mM KCl, and 1 mM TCEP.
Expression of the rat Munc13C (residues 529–1725, Δ1408–1452) was induced with 0.5 mM IPTG and performed overnight at 16°C. The protein was purified by affinity chromatography using HisPur Ni-NTA resin (Thermo Fisher Scientific) with extensive washes with 50 mM tris (pH 8), 10 mM imidazole, 750 mM NaCl, 1 mM TCEP, and 10% (v/v) glycerol. The protein was eluted with 50 mM tris (pH 8), 250 mM NaCl, 1 mM TCEP, 10% (v/v) glycerol, and 500 mM imidazole and dialyzed overnight at 4°C in 50 mM tris (pH 8), 250 mM NaCl, 1 mM TCEP, 2.5 mM CaCl2, and 10% (v/v) glycerol in the presence of thrombin. The protein was further purified by anion exchange chromatography on a HiTrap Q column (GE) in 20 mM tris (pH 8.0), 1 mM TCEP, and 10% (v/v) glycerol using a linear gradient from 0 to 1 M NaCl.
The following mutants were generated using the QuikChange site-directed mutagenesis and custom-designed primers and purified as the unmodified constructs: full-length syntaxin-1A (1–288) M138A; full-length syntaxin-1A (1–288) D184P; syntaxin-1A (2–253) M183A; syntaxin-1A (2–253) D184P; syntaxin-1A (2–253) C145A, L165A, E166A, L205C; syntaxin-1A (2–253) C145A, L165A, E166A, M183A, L205C; full-length syntaxin-1A (1–288) C145A, L165A, E166A, M183A, L205C; full-length syntaxin-1A (1–288) C145A, L165A, E166A, D184P, L205C; synaptobrevin-2 29–83 Q36C; Munc18-1 S42Q; Munc18-1 L307R; Munc18-1 Q301D; and Munc18-1 E352K.
Template complex formation
Before complex formation, TCEP was removed from each protein preparation using Superdex 75 10/300 GL column. TCEP-free Syb (100 to 200 μM) was mixed with 400 to 500 μM 2,2-dithiodipyridine (Sigma-Aldrich). The reaction was monitored at 343 nm using an Agilent 8453 ultraviolet-visible spectrometer. Upon completion, the excess of 2,2-dithiodipyridine was removed using a PD MiniTrap G-25 Sephadex column with 100 mM phosphate at pH 7.0, 100 mM NaCl, 6 M urea, and 0.2 mM EDTA as the elution buffer. An excess of activated Syb was then mixed with TCEP-free SyxLE and left incubated at room temperature overnight. The efficiency of disulfide bond formation between Syb and SyxLE was estimated on the basis of the absorbance at 343 nm. The SyxLE/Syb conjugate was purified by size exclusion chromatography using Superdex 75 10/300 GL and 20 mM Hepes (pH 7.4), 150 mM KCl as the running buffer. TCEP-free Munc18-1 D326K was then mixed with purified SyxLE/Syb and left rotated overnight at room temperature. The template complex was finally purified by size exclusion chromatography using a Superdex 75 10/300 GL column in 20 mM Hepes (pH 7.4) with 150 mM KCl.
EM data acquisition
Fluorinated fos-choline-8 was added into purified template complex formed by SyxLE and Munc18-1 D326K (7.5 mg/ml) to a final concentration of 5 mM, and 3 μl of the sample was applied to glow-discharged (30 mA, 80 s) Quantifoil R1.2/1.3 300-mesh gold holey carbon grids (Quantifoil, Micro Tools GmbH, Germany). Grids were blotted for 4.0 s under 100% humidity at 4°C before being plunged into the liquid ethane using Mark IV Vitrobot (FEI). Micrographs were acquired on a Titan Krios microscope (FEI) operated at 300 kV with a K3 direct electron detector (Gatan), using a slit width of 20 eV on a GIF-Quantum energy filter. SerialEM was used for data collection. A calibrated magnification of 46,296 was used for imaging of the samples, yielding a pixel size of 1.08 Å on the images. The defocus range was set from −1.6 to −2.6 μm. Each micrograph was dose-fractionated to 30 frames with a total dose of about 60 e−/Å2.
Image processing
The cryo-EM refinement statistics are summarized in table S1. A total of 7401 movie frames of the SyxLE/Syb–Munc18-1 D326K complex were motion-corrected, binned twofold, resulting in a pixel size of 1.08 Å, and dose-weighted using MotionCor2. The contrast transfer function (CTF) parameters were estimated using Gctf. RELION3 (53) was used for the following processing. Particles were first roughly picked by using the Laplacian-of-Gaussian blob method and then subjected to 2D classification. Class averages representing projections of the SyxLE/Syb-Munc18-1 D326 complex in different orientations were used as templates for reference-based particle picking. Extracted particles were binned three times and subjected to 2D classification. Particles from the classes with fine structural features were selected for 3D classification using an initial model generated from a subset of the particles in RELION3. Particles from one of the resulting 3D classes showing good secondary structural features were selected and reextracted into the original pixel size of 1.08 Å. Subsequently, we performed finer 3D classification imposed by using local search in combination with small angular sampling (3.75°), resulting in new classes showing two distinct conformations of the SyxLE/Syb–Munc18-1 D326 complex. The particles corresponding to two different conformations were selected and refined separately, leading to two different cryo-EM maps at 3.5-Å (class2) and 3.7-Å (class1) resolution, respectively.
Model building and refinement
Model building of both template complexes was started by docking the individual chains from the previously solved crystal structure of the syntaxin-1–Munc18-1 complex [Protein Data Bank (PDB) ID: 3C98] using Chimera 1.15 (54). The models were improved by iterative manual building in Coot 0.9 (55) and real-space refinement in the software package Phenix 1.19.1 (56). The density for Hd in syntaxin in both tc1 and tc2 is rather weak, preventing the assignment of individual residues. This part was therefore built as a polyalanine helix, with the register of the residues undetermined. The quality of the model stereochemistry and fit to density was evaluated with the comprehensive validation method MolProbity (57) as implemented in the Phenix package. The statistics are shown in table S1. The PyMOL Molecular Graphics System, Version 4.6.0 (Schrödinger LLC) was used for visualization of the structures, preparation of molecular diagrams for the figures, and calculations of buried surface areas.
Gel filtration–binding assay
Samples (6 to 20 μM) containing Munc18-1 D326K, Munc18-1 D326K incubated with SyxLE and Syb, or Munc18-1 D326K incubated with SyxLE/Syb were injected into a size exclusion chromatography column (Superdex 30 Increase 10/300 GL) using 20 mM Hepes (pH 7.4), 150 mM KCl as the running buffer. To form potential complexes, selected proteins were incubated overnight at 4°C before injection. Selected eluted fractions were loaded into SDS-PAGE gel and stained with InstantBlue Coomassie protein stain to confirm protein coelution.
SDS-PAGE SNARE complex assembly assay
To detect the SDS-resistant SNARE complex, the template complex was formed as described above, and the SyxLE/Munc18-1 D326K complex was preformed (overnight incubation at 4°C). SNAP-25 (5 μM) was added to 5 μM template complex; 5 μM Syb and 5 μM SNAP-25 were added to the SyxLE/Munc18-1 D326K complex; or 5 μM Syb, 5 μM SNAP-25, and 5 μM SyxLE were mixed. The reaction was done in 25 mM Hepes (pH 7.4), 150 mM KCl, and 10% (v/v) glycerol at room temperature. After 3 min, the reaction was stopped by addition of the SDS-PAGE gel-loading buffer. The samples were then loaded into SDS-PAGE gels and stained with InstantBlue Coomassie protein stain. The gels were imaged using a Bio-Rad ChemiDoc imaging system.
Mass photometry
Samples containing 1000 nM SyxLE/Syb, or SyxLE M183A/Syb, or SyxLE D184P/Syb, or syntaxin-1 (2–253), or syntaxin-1 (2–253) M183A, syntaxin-1 (2–253) D184P were incubated overnight at room temperate with TCEP-free 1000 nM or 2000 nM WT or mutant Munc18-1. Before the measurements, the samples were diluted 10-fold using PBS at pH 7.4. High-precision microscope cover glasses were rinsed with Milli-Q water, isopropanol, Milli-Q water, isopropanol, and Milli-Q water and dried using a stream of nitrogen gas. Clean coverslips with attached silicon gaskets were then mounted on immersion oil (refractive index of 1.518)–covered lenses of a Refeyn’s second-generation mass photometer. All the measurements were performed using PBS at pH 7.4. Data were collected using AcquireMP software. A single gasket was filled with 13 to 17.1 μl of PBS to enable focusing of the coverslip. Once the focus signal was stable, 0.9 to 7 μl of the 100 nM sample were added and quickly mixed to achieve the desired final protein concentrations. A movie was recorded for 60 s (2819 frames) and processed using DiscoverMP. Contrast-to-mass calibration was achieved using a bovine serum albumin (BSA) standard (0.002 mg/ml). The contrasts observed for BSA monomer (66 kDa), dimer (132 kDa), and trimer (198 kDa) were used to generate a standard calibration curve. Each measurement is displayed as normalized histograms with Gaussian fitting of the binding event counts with a loaded standard calibration curve. KD was calculated using a standard one-ligand binding model.
Solution SNARE complex assembly assay
To monitor SNARE complex assembly in solution, synaptobrevin (1–96) L26C and SNAP-25 R136C (50 to 150 μM) were respectively labeled with Alexa Fluor 488 maleimide and tetramethylrhodamine (TMR) maleimide at room temperature with 10- to 20-fold excess of the dyes for 2 hours. The excess of the reagents was removed using a Superdex 75 (10/300) column equilibrated with 20 mM tris (pH 7.4), 150 mM NaCl, and 1 mM TCEP. SNARE complex assembly was monitored by detecting Alexa Fluor 488 donor fluorescence intensity (excitation at 468 nm and emission at 518 nm) as a function of time at 37°C using a PTI Quantamaster 400 spectrofluorometer (T-format) equipped with a rapid Peltier temperature–controlled four-position sample holder with a GG495 long-pass filter mounted. To start the reactions, 0.1 μM synaptobrevin (1–96) L26C–Alexa Fluor 488 was mixed with 1 μM SNAP-25 R136C-TMR and 1 μM syntaxin-1 (2–253)/Munc18-1 complexes (preformed for 1 hour at room temperature). The reaction buffer was 25 mM Hepes (pH 7.4), 150 mM KCl, 1 mM TCEP, and 10% (v/v) glycerol.
Trans-SNARE complex formation assay
Single-cysteine variants of syntaxin-1A S186C and synaptobrevin-2 L26C were labeled with TMR and Alexa Fluor 488, respectively, as described above. V-liposomes were prepared similarly to those used for content mixing fusion assays (see below) except that they contained synaptobrevin-2 L26C–Alexa Fluor 488 with a 1:10,000 protein-to-lipid ratio. S-liposomes were prepared as the T-liposomes used for the content mixing assays but using syntaxin S186C-TMR without SNAP-25. Trans-SNARE complex formation was measured by the development of FRET between Alexa Fluor 488–synaptobrevin on V-liposomes (0.0625 mM total lipid) and TMR–syntaxin-1A on S-liposomes (0.25 mM total lipid) at 37°C using a PTI Quantamaster 400 spectrofluorometer (T-format) equipped with a rapid Peltier temperature–controlled four-position sample holder. Before the measurements, the S-liposomes were incubated with 0.37 μM WT or mutant Munc18-1 for 1 hour at room temperature. The Alexa Fluor 488 donor fluorescence of V-liposomes at 518 nm (excitation at 468 nm) was recorded to monitor the development of FRET over time upon mixing with S-liposomes containing the preformed syntaxin-1/Munc18-1 complexes, 2 μM SNAP-25a M71D, L78D, 0.2 μM Munc13C, and 0.1 mM EGTA. At 1100 s, the reaction was paused and 0.6 mM CaCl2 was quickly added to each reaction. The reaction buffer contained 25 mM Hepes (pH 7.4), 150 mM KCl, 1 mM TCEP, and 10% (v/v) glycerol. A GG495 long-pass filter (Edmund Optics) was used to filter scattered light.
Content mixing assays
The assay was performed as previously described (17). Briefly, V-liposomes containing full-length synaptobrevin-2 (protein-to-lipid ratio of 1:500) were prepared with 39% 1-palmitoyl, 2-oleoyl phosphatidylcholine (POPC), 19% 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), 19% 1-palmitoyl-2-oleoyl-sn-phosphatidylethanolamine (POPE), 20% cholesterol, 1.5% N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt (NBD-PE), and 1.5% 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (Marina Blue DHPE). T-liposomes containing syntaxin-1 (WT or M183, or D184P; syntaxin-1–to–lipid ratio of 1:800) and 25 μM SNAP-25 were prepared with 38% POPC, 18% DOPS, 20% POPE, 20% cholesterol, 2% phosphatidylinositol 4,5-biphosphate (PIP2), and 2% diacylglycerol (DAG). Dried lipid films were resuspended in 25 mM Hepes (pH 7.4), 150 mM KCl, 1 mM TCEP, and 2% octyl β-glucopyranoside (βOG). Lipid solutions were then mixed with the respective proteins and with 4 μM phycoerythrin-biotin for T-liposomes or with 8 μM Cy5-streptavidin for V-liposomes in 25 mM Hepes (pH 7.4), 150 mM KCl, 1 mM TCEP, and 10% (v/v) glycerol. Proteoliposomes were prepared by detergent removal using dialysis with Amberlite XAD-2 beads (2 g/liter; Sigma-Aldrich) three times at 4°C and subsequent co-floatation on a three-layer histodenz gradient (35, 25, and 0%) and harvested from the topmost layer. Content mixing signals were measured from the development of FRET between Cy5-streptavidin trapped in V-liposomes and phycoerythrin-biotin trapped in T-liposomes (excitation at 565 nm and emission at 670 nm). Each reaction was prepared in a total volume of 200 μl with V-liposomes (0.125 mM total lipid), T-liposomes (0.25 mM total lipids), 2.5 mM MgCl2, 2 mM ATP, 0.1 mM EGTA, 5 μM streptavidin, and the following proteins: 1 μM SNAP-25, 0.4 μM NSF purified in ATP-containing buffer, 2 μM αSNAP, 1 μM Munc18-1 (WT or other tested mutants), and 0.2 μM Munc13C. At 300 s, the reaction was paused and 0.6 mM CaCl2 was added to each reaction mixture and quickly mixed. All experiments were performed at 30°C using a PTI Quantamaster 400 spectrofluorometer (T-format) equipped with a rapid Peltier temperature–controlled four-position sample holder. Content mixing measurements were normalized to control reactions collected without streptavidin in the presence of 1% βOG to measure the maximal Cy5 fluorescence attainable.
Statistics
To analyze the statistical significance of the results obtained in various assays (Figs. 3 to 5), all experiments were performed at least in triplicates and one-way analysis of variance (ANOVA) was performed through an all-pairwise multiple comparison procedure using the Holm-Sidak test (***P < 0.001 and **P < 0.01). For the analysis of binding of WT and Munc18-1 mutants to SyxLE/Syb by mass photometry (Fig. 3I), the difference between the KD’s obtained for WT and S42Q mutant Munc18-1 was obviously statistically significant (P < 0.001), but the very large KD calculated for the S42Q mutant and the correspondingly large SD hinder the statistical analysis of significance for other pairwise comparisons within the group. Hence, all other comparisons within the group were performed after removing the S42Q data.
Acknowledgments
We thank S. Sugita for fruitful discussions and C. Brautigam for assistance with the mass photometry experiments.
Funding: Some data presented in this report were acquired with a mass photometer that was supported by award S10OD030312-01 from the NIH. This work was supported by Welch Foundation grants I-1304 (to J.R.), I-1944 (to X.-C.B.), and I-1702 (to X.Z.); by NIH Research Program Award R35 NS097333 (to J.R.); and by NIH Research Award R35GM130289 (to X.Z.). Cryo-EM data were collected at the University of Texas Southwestern Medical Center (UTSW) Cryo-Electron Microscopy Facility, funded in part by the Cancer Prevention and Research Institute of Texas (CPRIT) Core Facility Support Award RP170644.
Author contributions: K.P.S., X.-C.B., and J.R. conceived the research, K.P.S. conducted most experiments. K.P.S. and J.R. analyzed the data. J.X. froze the cryo-EM grids. X.-C.B. and J.X. acquired the cryo-EM data. X.-C.B. processed and analyzed the cryo-EM data. K.P.S. and X.Z. built and refined the structural models. J.R. and K.P.S. wrote the draft, with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The class1 and class2 cryo-EM structures have been deposited in the Protein Data Bank with accession numbers 7UDC and 7UDB, respectively.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Tables S1 to S3
REFERENCES AND NOTES
- 1.Sollner T., Bennett M. K., Whiteheart S. W., Scheller R. H., Rothman J. E., A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75, 409–418 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Poirier M. A., Xiao W., Macosko J. C., Chan C., Shin Y. K., Bennett M. K., The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat. Struct. Biol. 5, 765–769 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T., Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Hanson P. I., Roth R., Morisaki H., Jahn R., Heuser J. E., Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90, 523–535 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Rizo J., Molecular Mechanisms Underlying Neurotransmitter Release. Annu. Rev. Biophys. 51, 377–408 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez I., Ubach J., Dulubova I., Zhang X., Südhof T. C., Rizo J., Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell 94, 841–849 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Dulubova I., Sugita S., Hill S., Hosaka M., Fernandez I., Südhof T. C., Rizo J., A conformational switch in syntaxin during exocytosis: Role of munc18. EMBO J. 18, 4372–4382 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misura K. M., Scheller R. H., Weis W. I., Three-dimensional structure of the neuronal-Sec1–syntaxin 1a complex. Nature 404, 355–362 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Ma C., Su L., Seven A. B., Xu Y., Rizo J., Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prinslow E. A., Stepien K. P., Pan Y. Z., Xu J., Rizo J., Multiple factors maintain assembled trans-SNARE complexes in the presence of NSF and αSNAP. eLife 8, e38880 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma C., Li W., Xu Y., Rizo J., Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat. Struct. Mol. Biol. 18, 542–549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X., Wang S., Sheng Y., Zhang M., Zou W., Wu L., Kang L., Rizo J., Zhang R., Xu T., Ma C., Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat. Struct. Mol. Biol. 22, 547–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X., Seven A. B., Camacho M., Esser V., Xu J., Trimbuch T., Quade B., Su L., Ma C., Rosenmund C., Rizo J., Functional synergy between the Munc13 C-terminal C1 and C2 domains. eLife 5, e13696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quade B., Camacho M., Zhao X., Orlando M., Trimbuch T., Xu J., Li W., Nicastro D., Rosenmund C., Rizo J., Membrane bridging by Munc13-1 is crucial for neurotransmitter release. eLife 8, e42806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richmond J. E., Weimer R. M., Jorgensen E. M., An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature 412, 338–341 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S., Bin N. R., Yu B., Wong R., Sitarska E., Sugita K., Ma K., Xu J., Tien C. W., Algouneh A., Turlova E., Wang S., Siriya P., Shahid W., Kalia L., Feng Z. P., Monnier P. P., Sun H. S., Zhen M., Gao S., Rizo J., Sugita S., UNC-18 and tomosyn antagonistically control synaptic vesicle priming downstream of UNC-13 in Caenorhabditis elegans. J. Neurosci. 37, 8797–8815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stepien K. P., Prinslow E. A., Rizo J., Munc18-1 is crucial to overcome the inhibition of synaptic vesicle fusion by αSNAP. Nat. Commun. 10, 4326 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verhage M., Maia A. S., Plomp J. J., Brussaard A. B., Heeroma J. H., Vermeer H., Toonen R. F., Hammer R. E., van den T. K., Berg, Missler M., Geuze H. J., Südhof T. C., Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science 287, 864–869 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Richmond J. E., Davis W. S., Jorgensen E. M., UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat. Neurosci. 2, 959–964 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varoqueaux F., Sigler A., Rhee J. S., Brose N., Enk C., Reim K., Rosenmund C., Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming. Proc. Natl. Acad. Sci. U.S.A. 99, 9037–9042 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aravamudan B., Fergestad T., Davis W. S., Rodesch C. K., Broadie K., Drosophila UNC-13 is essential for synaptic transmission. Nat. Neurosci. 2, 965–971 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Koushika S. P., Richmond J. E., Hadwiger G., Weimer R. M., Jorgensen E. M., Nonet M. L., A post-docking role for active zone protein Rim. Nat. Neurosci. 4, 997–1005 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tien C. W., Yu B., Huang M., Stepien K. P., Sugita K., Xie X., Han L., Monnier P. P., Zhen M., Rizo J., Gao S., Sugita S., Open syntaxin overcomes exocytosis defects of diverse mutants in C. elegans. Nat. Commun. 11, 5516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parisotto D., Pfau M., Scheutzow A., Wild K., Mayer M. P., Malsam J., Sinning I., Söllner T. H., An extended helical conformation in domain 3a of Munc18-1 provides a template for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex assembly. J. Biol. Chem. 289, 9639–9650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munch A. S., Kedar G. H., van Weering J. R. T., Vazquez-Sanchez S., He E., Andre T., Braun T., Sollner T. H., Verhage M., Sorensen J. B., Extension of helix 12 in Munc18-1 induces vesicle priming. J. Neurosci. 36, 6881–6891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han G. A., Park S., Bin N. R., Jung C. H., Kim B., Chandrasegaram P., Matsuda M., Riadi I., Han L., Sugita S., A pivotal role for pro-335 in balancing the dual functions of Munc18-1 domain-3a in regulated exocytosis. J. Biol. Chem. 289, 33617–33628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cote M., Ménager M. M., Burgess A., Mahlaoui N., Picard C., Schaffner C., Al-Manjomi F., Al-Harbi M., Alangari A., Deist F. L., Gennery A. R., Prince N., Cariou A., Nitschke P., Blank U., El-Ghazali G., Ménasché G., Latour S., Fischer A., de Saint Basile G., Munc18-2 deficiency causes familial hemophagocytic lymphohistiocytosis type 5 and impairs cytotoxic granule exocytosis in patient NK cells. J. Clin. Invest. 119, 3765–3773 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryant N. J., Gould G. W., SNARE proteins underpin insulin-regulated GLUT4 traffic. Traffic 12, 657–664 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Xu Y., Su L., Rizo J., Binding of Munc18-1 to synaptobrevin and to the SNARE four-helix bundle. Biochemistry 49, 1568–1576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker R. W., Jeffrey P. D., Zick M., Phillips B. P., Wickner W. T., Hughson F. M., A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sitarska E., Xu J., Park S., Liu X., Quade B., Stepien K., Sugita K., Brautigam C. A., Sugita S., Rizo J., Autoinhibition of Munc18-1 modulates synaptobrevin binding and helps to enable Munc13-dependent regulation of membrane fusion. eLife 6, e24278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao J., He M., Port S. A., Baker R. W., Xu Y., Qu H., Xiong Y., Wang Y., Jin H., Eisemann T. J., Hughson F. M., Zhang Y., Munc18-1 catalyzes neuronal SNARE assembly by templating SNARE association. eLife 7, e41771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L., Rebane A. A., Yang G., Xi Z., Kang Y., Gao Y., Zhang Y., Munc18-1-regulated stage-wise SNARE assembly underlying synaptic exocytosis. eLife 4, e09580 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burkhardt P., Hattendorf D. A., Weis W. I., Fasshauer D., Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 27, 923–933 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisemann T. J., Allen F., Lau K., Shimamura G. R., Jeffrey P. D., Hughson F. M., The Sec1/Munc18 protein Vps45 holds the Qa-SNARE Tlg2 in an open conformation. eLife 9, e60724 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., Gong J., Zhu L., Wang S., Yang X., Xu Y., Yang X., Ma C., Munc13 activates the Munc18-1/syntaxin-1 complex and enables Munc18-1 to prime SNARE assembly. EMBO J. 39, e103631 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andre T., Classen J., Brenner P., Betts M. J., Dörr B., Kreye S., Zuidinga B., Meijer M., Russell R. B., Verhage M., Söllner T. H., The interaction of Munc18-1 Helix 11 and 12 with the central region of the VAMP2 SNARE motif is essential for SNARE templating and synaptic transmission. eNeuro 7, ENEURO.0278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young G., Hundt N., Cole D., Fineberg A., Andrecka J., Tyler A., Olerinyova A., Ansari A., Marklund E. G., Collier M. P., Chandler S. A., Tkachenko O., Allen J., Crispin M., Billington N., Takagi Y., Sellers J. R., Eichmann C., Selenko P., Frey L., Riek R., Galpin M. R., Struwe W. B., Benesch J. L. P., Kukura P., Quantitative mass imaging of single biological macromolecules. Science 360, 423–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colbert K. N., Hattendorf D. A., Weiss T. M., Burkhardt P., Fasshauer D., Weis W. I., Syntaxin1a variants lacking an N-peptide or bearing the LE mutation bind to Munc18a in a closed conformation. Proc. Natl. Acad. Sci. U.S.A. 110, 12637–12642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camacho M., Quade B., Trimbuch T., Xu J., Sari L., Rizo J., Rosenmund C., Control of neurotransmitter release by two distinct membrane-binding faces of the Munc13-1 C1C2B region. eLife 10, e72030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J., Camacho M., Xu Y., Esser V., Liu X., Trimbuch T., Pan Y. Z., Ma C., Tomchick D. R., Rosenmund C., Rizo J., Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN. eLife 6, e22567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song H., Wickner W., A short region upstream of the yeast vacuolar Qa-SNARE heptad-repeats promotes membrane fusion through enhanced SNARE complex assembly. Mol. Biol. Cell 28, 2282–2289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torng T., Song H., Wickner W., Asymmetric Rab activation of vacuolar HOPS to catalyze SNARE complex assembly. Mol. Biol. Cell 31, 1060–1068 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torng T., Wickner W., Phosphatidylinositol and phosphatidylinositol-3-phosphate activate HOPS to catalyze SNARE assembly, allowing small headgroup lipids to support the terminal steps of membrane fusion. Mol. Biol. Cell 32, ar19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dulubova I., Yamaguchi T., Wang Y., Sudhof T. C., Rizo J., Vam3p structure reveals conserved and divergent properties of syntaxins. Nat. Struct. Biol. 8, 258–264 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Wang S., Choi U. B., Gong J., Yang X., Li Y., Wang A. L., Yang X., Brunger A. T., Ma C., Conformational change of syntaxin linker region induced by Munc13s initiates SNARE complex formation in synaptic exocytosis. EMBO J. 36, 816–829 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magdziarek M., Bolembach A. A., Stepien K. P., Quade B., Liu X., Rizo J., Re-examining how Munc13-1 facilitates opening of syntaxin-1. Protein Sci. 29, 1440–1458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meijer M., Dörr B., Lammertse H. C., Blithikioti C., van Weering J. R., Toonen R. F., Söllner T. H., Verhage M., Tyrosine phosphorylation of Munc18-1 inhibits synaptic transmission by preventing SNARE assembly. EMBO J. 37, 300–320 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sudhof T. C., Neurotransmitter release: The last millisecond in the life of a synaptic vesicle. Neuron 80, 675–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X., Araç D., Wang T. M., Gilpin C. J., Zimmerberg J., Rizo J., SNARE-mediated lipid mixing depends on the physical state of the vesicles. Biophys. J. 90, 2062–2074 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X., Tomchick D. R., Kovrigin E., Araç D., Machius M., Südhof T. C., Rizo J., Three-dimensional structure of the complexin/SNARE complex. Neuron 33, 397–409 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Liu X., Seven A. B., Xu J., Esser V., Su L., Ma C., Rizo J., Simultaneous lipid and content mixing assays for in vitro reconstitution studies of synaptic vesicle fusion. Nat. Protoc. 12, 2014–2028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L. W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1 to S3