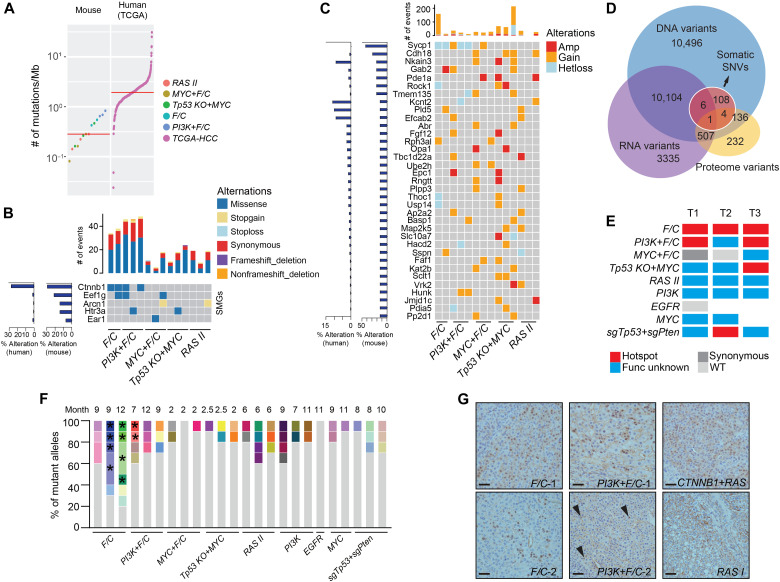
Fig. 4. Mutational profiles of mouse liver tumors.
(A) Mutational burden of mouse liver tumors and human HCC (the TCGA cohort, 358 cases) by WES. Red line indicates the median. (B and C) SMGs (B) and genes with CNV (C) in mouse tumors. Alteration frequencies in mouse tumors are indicated on the left in bar graphs. Numbers of events found in individual tumors are indicated on top. (D) Overlap of nonsynonymous SNVs and single–amino acid variants detected in WES, RNA-seq, and LC-MS/MS. (E) Hotspot mutations of Ctnnb1 exon 3 in specific genotypes. Exon 3 of Ctnnb1 was cloned and sequenced for one to three tumors in each of the nine genotypes, and the presence of hotspot mutations is indicated in red. (F) Diversity of Ctnnb1 alleles in mouse tumors. Different alleles are indicated by color. Alleles from a single tumor sharing a common mutation are indicated by asterisks. (G) Nuclear localization of Ctnnb1 in mouse tumors. Two tumors for each of F/C and PI3K+F/C genotypes are presented. RAS I was a negative control, and CTNNB1+RAS was a positive control. Arrowheads point to nuclear Ctnnb1 in PI3K+F/C tumor 2. Scale bars, 40 μm.