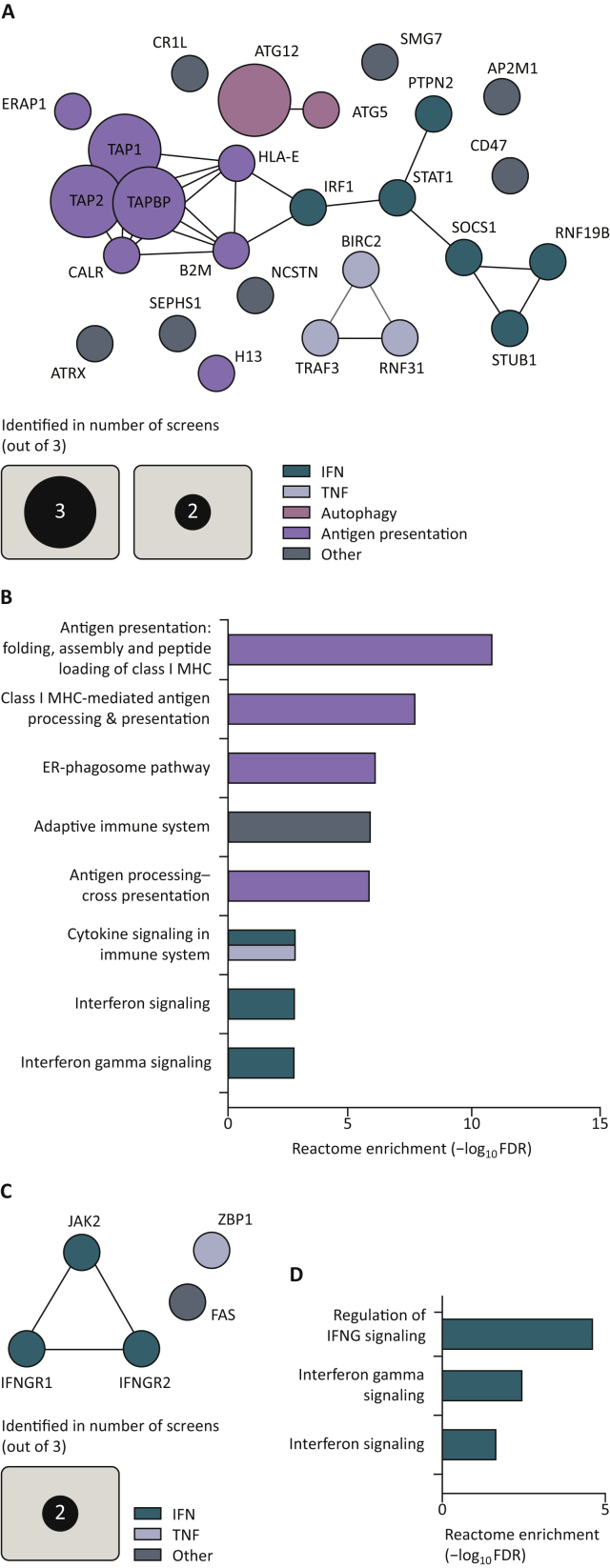
Figure 3.
In vivo screens identify common sensitivity hits and common resistance hits.
(A) Clustering analysis of significant in vivo sensitivity hits identified in at least two screens. Significance was determined by the original authors as P < 0.05 or FDR < 0.05. Murine gene symbols were translated to their human paralog by SynGO.149 Clustering was carried out by STRING.148 (B) Gene ontology enrichment scores of the eight top Reactome gene sets by Panther,150 based on in vivo sensitivity hits identified in at least two screens. Gene sets larger than 500 genes were excluded. (C) Clustering analysis of significant in vivo resistance hits identified in at least two screens. Significance was determined by the original authors as P < 0.05 or FDR < 0.05. Murine gene symbols were translated to their human paralog by SynGO.149 Clustering was carried out by STRING.148 (D) Gene ontology enrichment scores of the only significant Reactome gene sets by Panther,150 based on in vivo sensitivity hits identified in at least two screens. Gene sets larger than 500 genes were excluded.
ER, endoplasmic reticulum; FDR, false discovery rate; IFN, interferon; IFNG, interferon-γ; MHC, major histocompatibility complex; TNF, tumor necrosis factor.