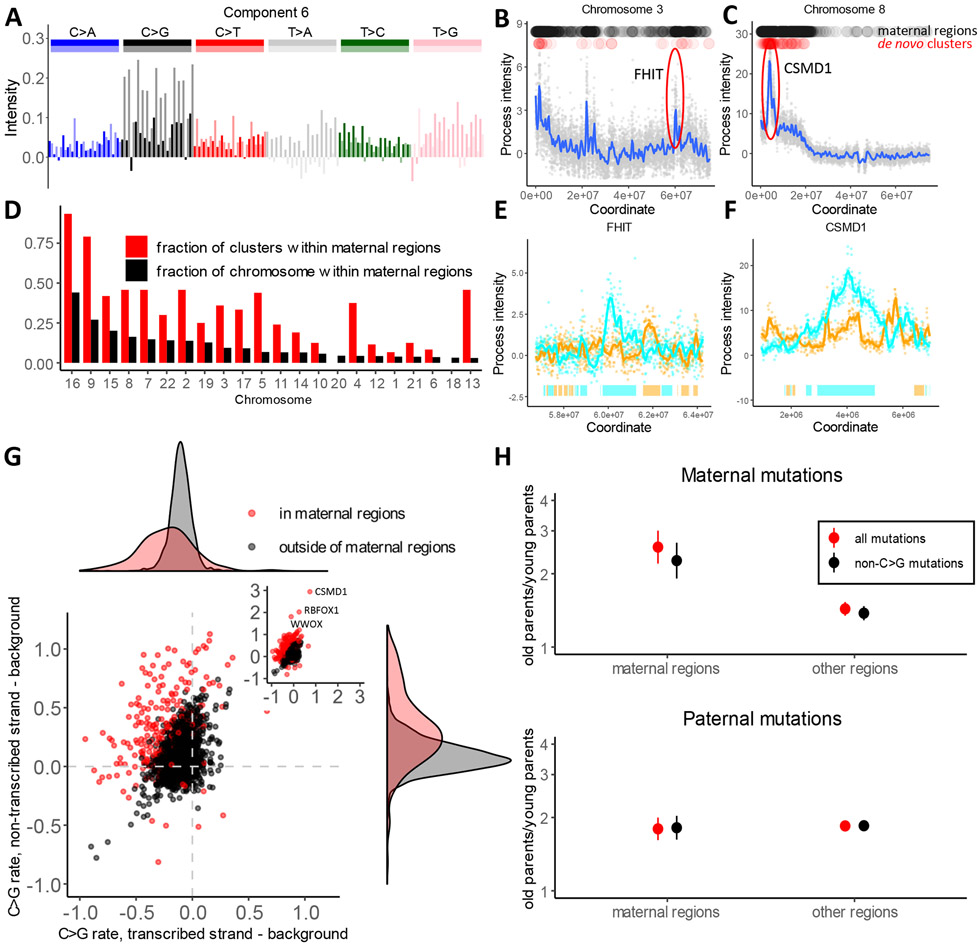
Fig 3. Oocyte-specific mutational process.
(A) The spectrum of one of the two components comprising process 6/7.
(B and C) Examples of two loci with high intensity of process 6/7, estimated as the sum of intensities of component 6 and component 7. Black dots on top of the panels mark windows of high intensity that we call “maternal regions” (see Methods). Red dots show de novo maternal clustered mutations from Halldorsson et al. (18).
(D) Enrichment of maternal clustered de novo mutations from Halldorsson et al. in maternal regions. The fraction of each chromosome that is attributed to “maternal regions” (high intensity of process 6/7) is shown in black. The fraction of maternal clustered mutations located within maternal regions on each chromosome is shown in red; the difference in size between the red and black bars indicates enrichment of clustered mutations within “maternal regions”.
(E and F) Zoom in view of process 6/7 intensity spikes around FHIT and CSMD1 genes on non-transcribed strands. Bars on the bottom depict gene bodies (colors: cyan if transcribed strand is the reference strand and orange otherwise).
(G) Difference between C>G mutation rate on transcribed or non-transcribed strand of a gene compared to a 100 KB region flanking the gene. Red dots correspond to genes within maternal regions and black dots corresponds to genes outside of maternal regions. Density plots on the top and right summarize the distributions on the X and Y axes.
(H) Ratio of parent-specific de novo mutation rates between first and last parent age quartiles is shown, estimated independently for “maternal regions” and for the rest of the genome. The error bars show the 95% confidence interval (95% CI) for the ratio of two binomial proportions test.