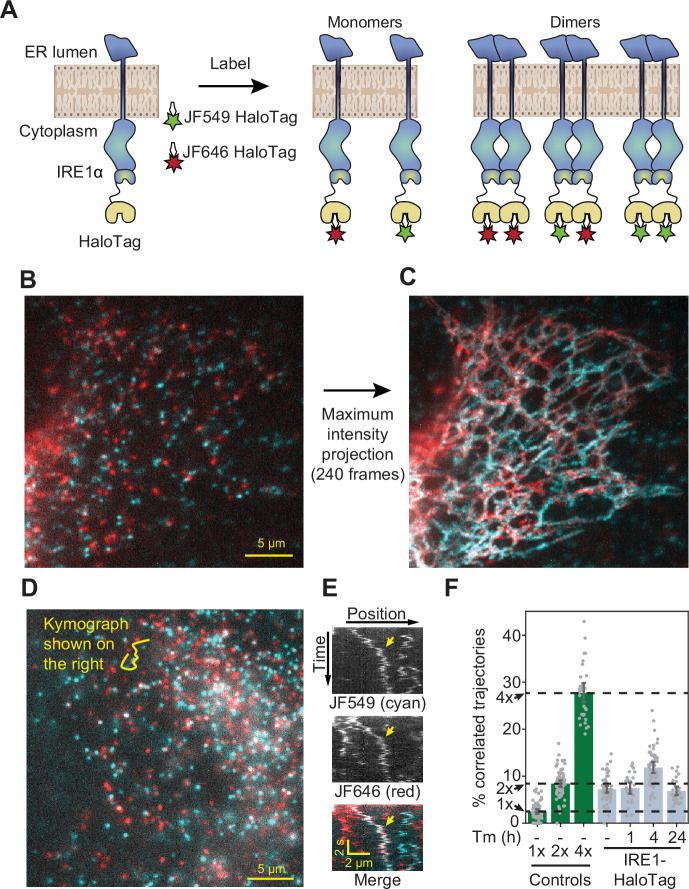
Figure 3. Detection of IRE1 dimers and oligomers in live cells.
(A) Schematic depiction of the assay. IRE1-HaloTag is simultaneously labeled with HaloTag dyes of two different colors, JF549 and JF646. If the protein is purely monomeric, all single-molecule tracks are expected to be either one color or the other. If it is purely dimeric, a fraction of tracks will contain both colors. Such dual-color tracks can then be identified as correlated trajectories. (B) Single frame from a long-exposure movie (100ms per frame) of a cell in which IRE1-HaloTag is labeled with a mixture of JF549 (cyan) and JF646 (red) dyes. (C) Maximum intensity projection of the entire movie from panel B showing that single IRE1 molecules diffuse along ER tubules. (D) Single frame from a short-exposure movie (50ms per frame) of a cell in which IRE1-HaloTag is labeled with a mixture of JF549 (cyan) and JF646 (red) dyes. (E) Kymograph (time vs. position plot) along the line shown in panel D. Co-localizing diffusional IRE1 trajectory is shown with a yellow arrow. (F) Stress-induced changes in IRE1 oligomerization in response to treatment with 5 μg/ml tunicamycin (Tm), as quantified by the fraction of correlated trajectories. Green bars on the left correspond to the 1 x, 2 x, and 4 x HaloTag controls, respectively. Error bars represent 95% confidence intervals.
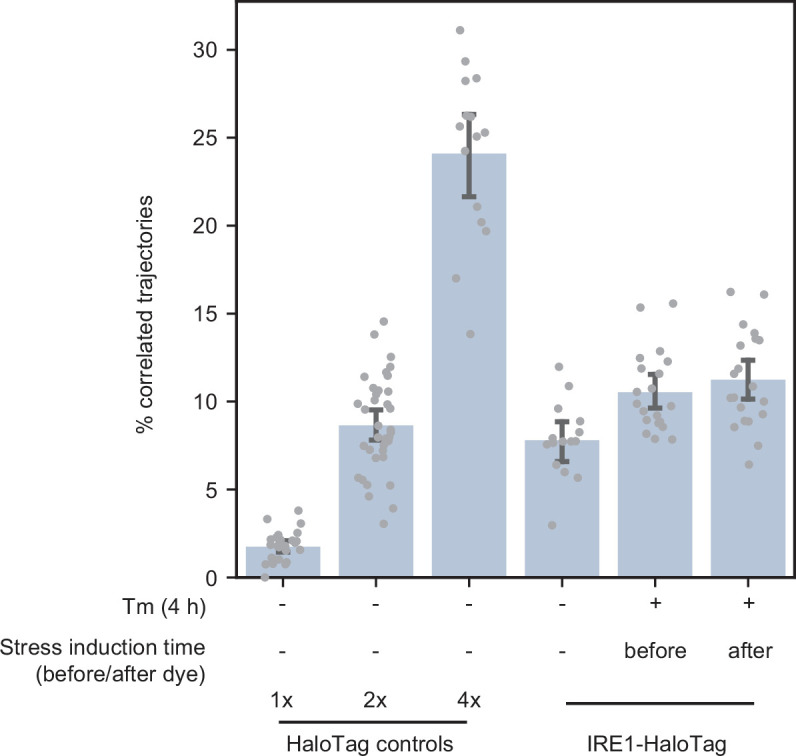
Figure 3—figure supplement 1. Effect of ER stress on HaloTag controls.
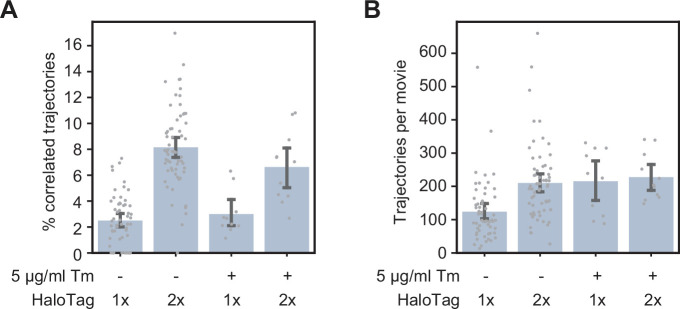
Figure 3—figure supplement 2. Effect of ER stress on the efficiency of HaloTag labeling.
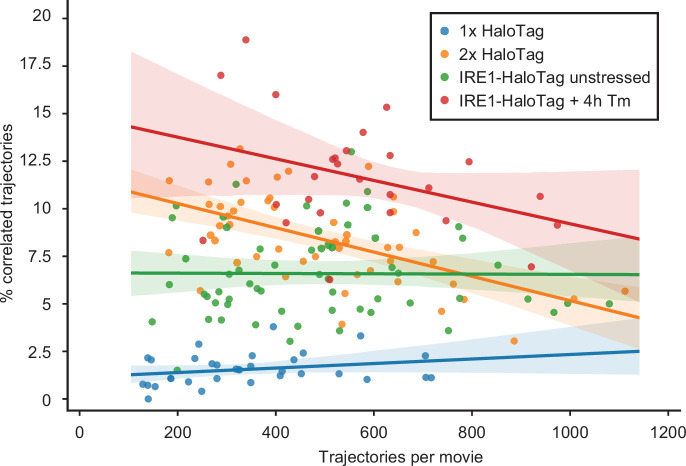
Figure 3—figure supplement 3. Quantification of diffusion from single-particle trajectories.