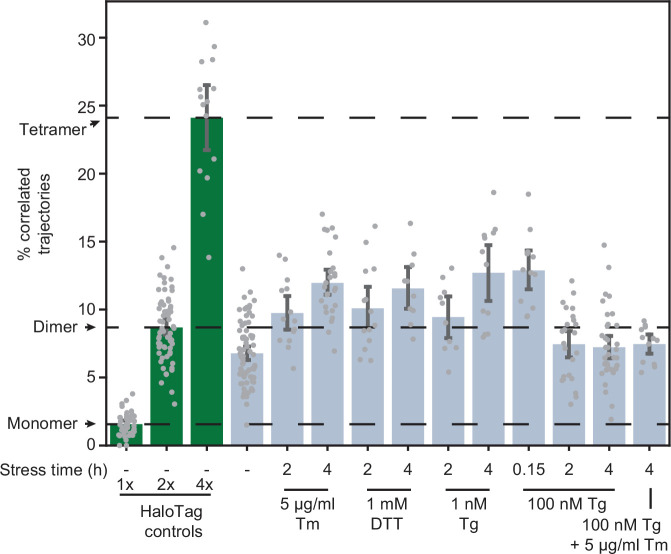
Figure 4. Effects of stressors on IRE1 oligomerization.
Oligomerization of endogenously tagged IRE1-HaloTag in U-2 OS cells treated with the indicated ER stressors for the indicated amounts of time. Tunicamycin (Tm) inhibits glycosylation in the ER lumen, thapsigargin (Tg) blocks sarco/endoplasmic reticulum Ca2+ pumps, and dithiothreitol (DTT) triggers reduction of disulfide bonds. Green bars on the left correspond to the 1 x, 2 x, and 4 x HaloTag controls, respectively. Error bars represent 95% confidence intervals.
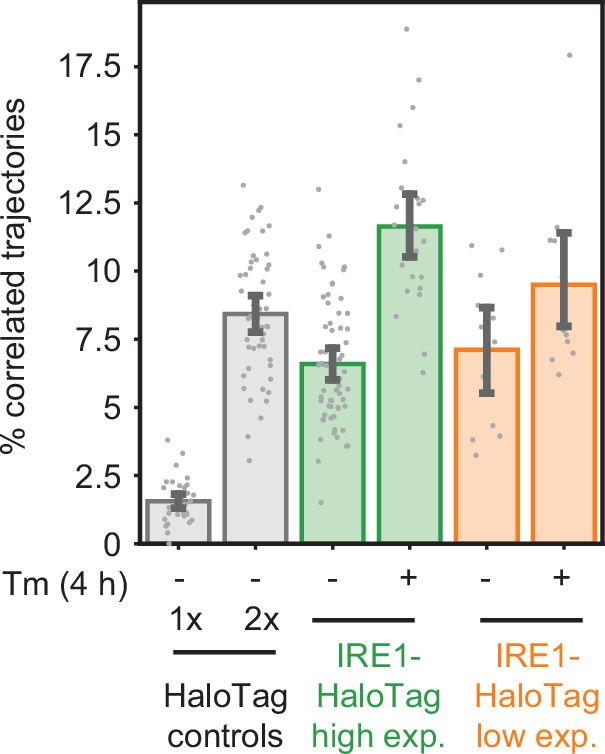
Figure 4—figure supplement 1. Formation of dimers and oligomers in high- and low-expressing clones of IRE1-HaloTag cells.
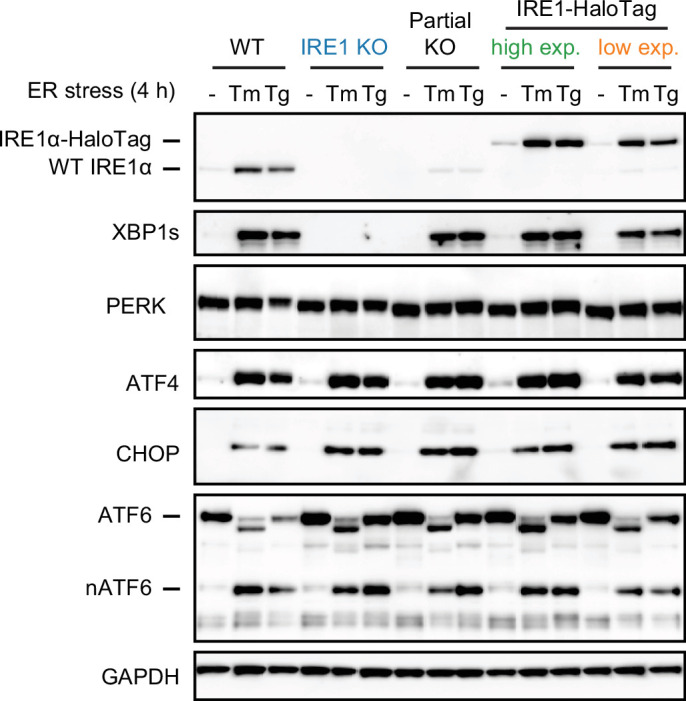
Figure 4—figure supplement 2. Trajectory density vs. percent correlation.