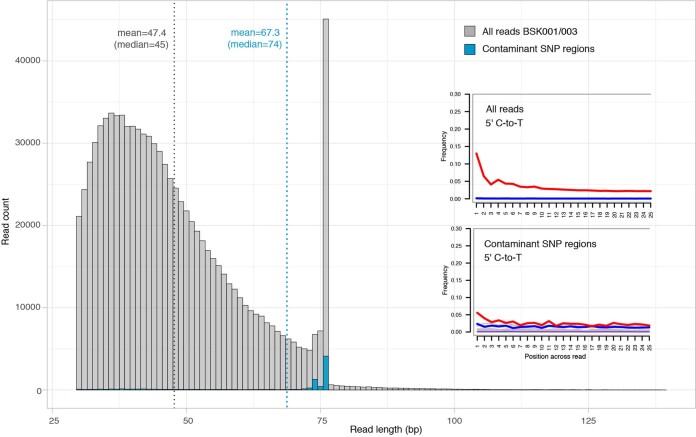
Extended Data Fig. 4. Read length and ancient DNA damage distribution of contaminant SNP regions in the combined BSK001/003 dataset prior to metagenomic filtering.
Overlayed length distributions of reads mapping against the CO92 Y. pestis reference genome, calculated for the entire dataset (gray) as well as for 117 regions surrounding putatively contaminant SNPs (blue). Regions were extracted within a 150 bp window surrounding each putatively contaminant SNP. Dotted lines represent average fragment lengths for the entire dataset and for the 117 putatively contaminant SNP regions in gray and blue, respectively. Reads comprising contaminant SNP regions show a distinct length distribution compared to the one observed across the entire BSK001/003 genome, with a marked shift towards longer read lengths. The 76 bp fragment length peak represents the uppermost possible read length of single-end sequenced reads, which comprised the majority of data within the present dataset. Ancient DNA damage patterns were compared between the entire dataset (upper panel) and the putatively contaminant SNP regions (lower panel), showing a near 3-fold reduction in the latter as estimated for the terminal 5′ base.