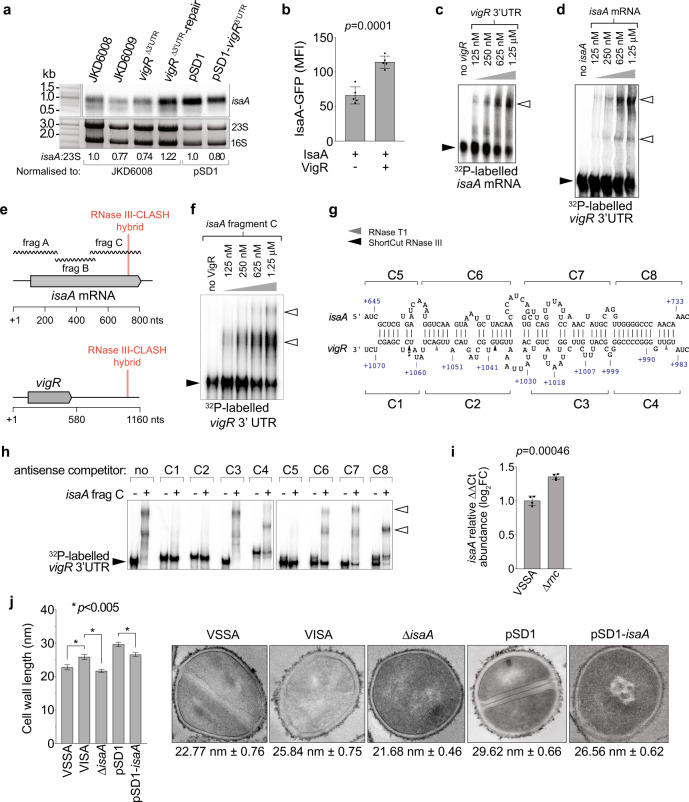
Fig. 6. The lytic transglycosylase IsaA is regulated by the 3′UTR of vigR.
a Northern analysis of isaA abundance in VISA (JKD6008) and isogenic vigR∆3’UTR, vigR∆3’UTR-repair, along with JKD6008 pSD1 and pSD1-vigR 3′UTR. Sybr Green II stained 23S and 16S rRNA are shown as loading controls. Quantification of the ratio of 23S rRNA to isaA by densitometry is indicated below. b Quantification of expression of a constitutively transcribed IsaA-GFP fusion (pCN33::isaA-gfp) with or without expression of vigR 3′UTR (pICS3::vigR) (indicated below). Median fluorescence intensity (MFI) is reported on the y-axis. Histogram heights represents the mean MFI and error bars indicate standard error from n = 5 biological replicates. The p-value is calculated using a two-sided t-test. c EMSA analysis of interactions between vigR 3′UTR and full-length isaA mRNA. 50 fM of radiolabelled isaA mRNA was titrated against increasing concentrations of vigR 3′UTR (top). d 50 fmol of radiolabelled vigR 3’UTR was titrated against increasing concentrations of full-length isaA mRNA. e Schematic of the RNase III-CLASH hybrid read within isaA mRNA and vigR mRNA. The isaA mRNA was synthesised as sub-fragments (frag A–C, wavy lines) ~300-nt in length to be used for EMSA. f isaA fragment C (frag-C). Concentrations of isaA are indicated (top). Black arrowheads indicated migration of free, radiolabelled RNA and open arrowheads indicate slow migrating vigR-isaA duplexes. g Predicted interaction between RNAs within the isaA-vigR hybrid read. The start and end positions of the RNA–RNA duplex are indicated respresentative of the mRNA TSS. The antisense oligonucleotide competitors used for EMSAs are detailed above and below their respective mRNAs (C1–C83). The cleavage sites of RNase T1 and ShortCut RNase III detected by primer extension are indicated by the grey and black arrows, respectively (also see Supplementary Fig. 6C, D). h EMSA analysis of interactions between vigR 3′UTR and isaA frag-C (0 or 1.25 μM concentration). Black arrowheads indicate migration of free radiolabelled vigR 3′UTR RNA. Antisense competitor oligonucleotides C1–C8 (indicated on top) were spiked in at 500× excess concentration. Open arrowhead indicates slow migrating vigR-isaA frag-C duplexes. I Quantitative RT-PCR for isaA mRNA abundance (relative to gapA) in VSSA and ∆rnc strain grown in BHI to an OD 578 nm of 3.0. Histogram height represents the mean and error bars indicate the standard deviation (SD) from n = 3 biological replicates. The p-value is calculated using a two-sided t-test. J Histogram of cell wall thickness for VSSA, VISA, and isaA deletion and knockdown strains (left). Error bars represent standard error of the mean (SEM) from n = 100 measurements. The p-value is calculated using a two-sided t-test. *p < 0.005. (right) Representative transmission electron microscopy (TEM) images of VSSA (JKD6009), VISA (JKD6008), and JKD6008 derivatives. The average cell wall thickness and standard error are shown below.