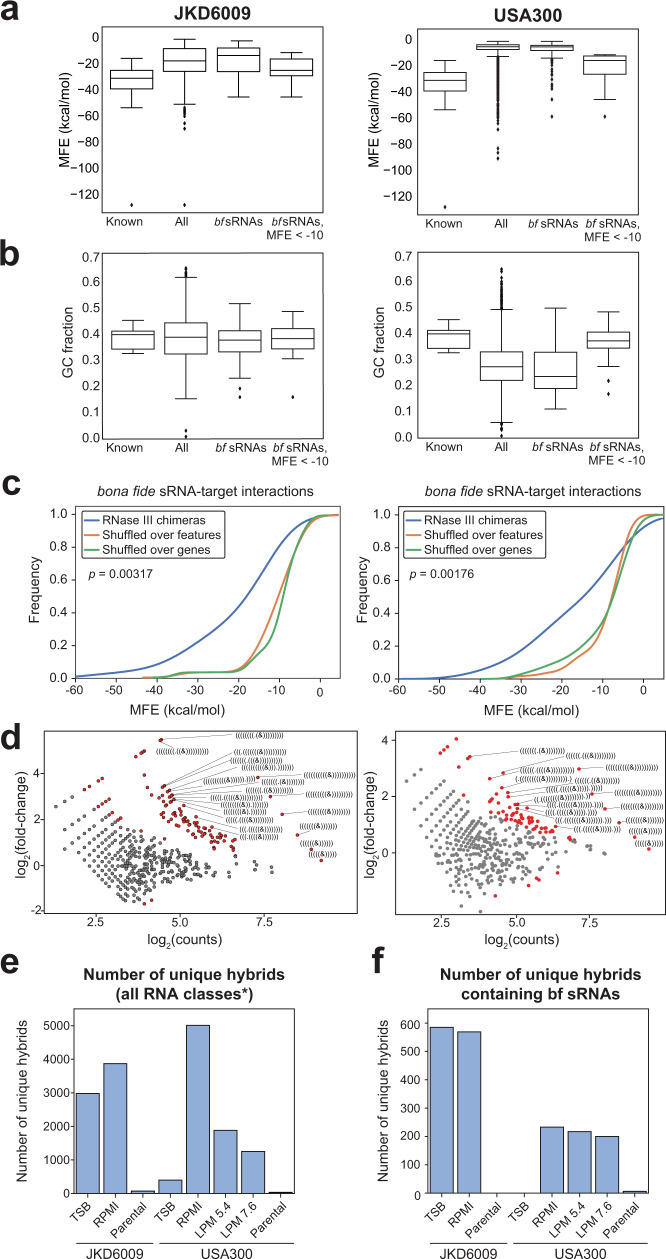
Fig. 2. Folding and structural analyses of hybrids identified through RNase III CLASH.
a Boxplots showing the minimum folding energy (MFE) of identified RNA-RNA interactions in all independent RNase III CLASH datasets (n = 4). Plotted are the previously verified interactions (“Known”) captured through CLASH, all the interactions identified by CLASH; only those containing a bona fide sRNA (bf sRNAs); and those containing a bona fide sRNA and filtered for MFE < -10 kcal/mol. The boxplot extends from first to the third quartile values of the data, with a line at the median. The whiskers extend from the edges of box to show the interquartile range multiplied by 1.5. Outliers are plotted as separate dots. b As in a but with reference to GC content. c Cumulative distribution of the MFE of the filtered interactions involving bona fide sRNAs in JKD6009 (left) and USA300 (right). Folding energies were calculated using RNADuplex53. As controls, interactions were shuffled randomly against other partners of the same class (orange line) or randomly across the gene (green line) of their partner identified through CLASH. Significance was tested with the Kolmgorov-Smirnov test. d Enriched structural motifs (red dots) in hybrids identified through RNase III CLASH. The incidence of each structure generated by RNADuplex was counted and compared to interactions randomly shuffled against different partners. Significance was calculated using a one-sided Fisher’s exact test and Benjamini-Hochberg correction was applied to account for multiple tests. The x-axis displays the total number of counts for each structure in the data, whereas the y-axis indicates the log2-fold difference for each structure between the experimental data and the randomly shuffled data. e Total number of unique hybrids identified in each experimental condition. The parental controls for each strain were merged. *tRNA-tRNA and rRNA-rRNA chimeras were excluded due to their high sequence similarity, meaning that we could not unambiguously determine if these represented intermolecular or intramolecular interactions. f As in e, but with respect to hybrids involving a bf sRNA.