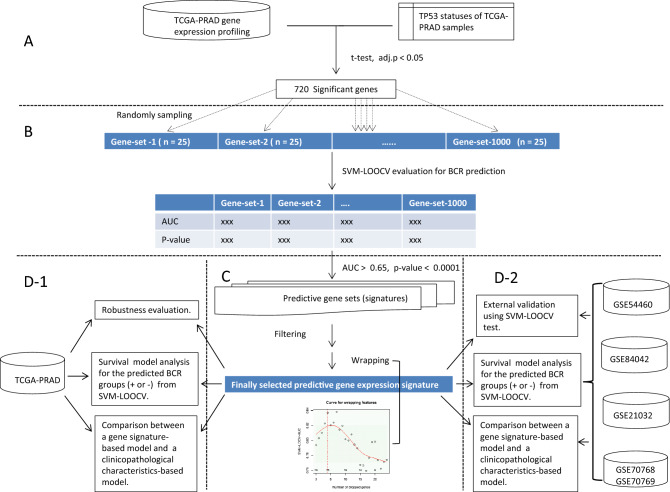
Figure 1.
Flow chart of the identification approach (A,B,C) and performance/utility evaluation (D-1, D-2) of a TP53 mutation status-associated predictive transcriptomic signature for BCR. In A, the top differentially expressed genes (DEGs) regarding TP53 statuses were identified. In (B), 1000 small subsets of the DEGs were randomly sampled and their predictive strengths for BCR were assessed by SVM-based cross validation. In (C), the results from (B) step were integrated by a “filter” and a novel “wrapper” to obtain an optimized gene signature. In (D1) and (D2), the performance of the finally selected signature for BCR prediction and the clinical utility were evaluated in the TCGA dataset and five external datasets using statistical and machine learning methods. See the main text for a more detailed explanation.