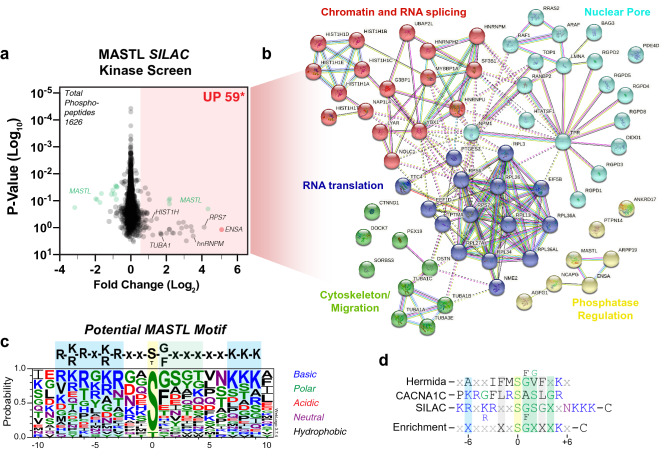
Figure 2.
MASTL SILAC kinase screen. (a) Volcano plot of all identified phospho-peptides identified from the MASTL SILAC kinase screen. A minimal Log2 Fold change cut-off < 0.5 (red shading) was taken as potential increased phosphorylation events. (b) STRING49 cluster analysis of proteins corresponding to increased phospho-peptides identified in a. (c) Phospho-peptides with a Log2 Fold change > 1.0 were aligned using WebLogo v3.7.452. Residue colors indicate chemical properties of amino acids. (d) Alignment of top phosphorylated peptide (CACNA1C) from the S/T/Y Physiological Kinase Substrate Finder Assay (Reaction Biology), with the potential MASTL motif identified in c.