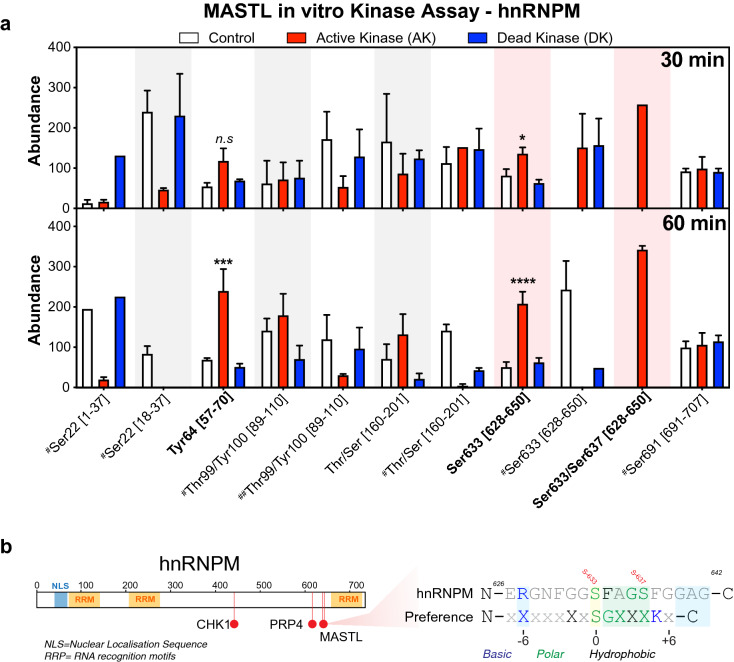
Figure 4.
In vitro kinase assay validation of hnRNPM phosphorylation by MASTL. (a) The ion abundance of each phospho-site activated in triplicate reactions between MASTL kinase and hnRNPM (as detailed in this figure legend). Phospho-site abundances were calculated using label-free quantitation. Square brackets indicate peptide locations within the protein on which each phospho-site is found. Single hashtags denote a co-modification within that peptide, e.g., carbamidomethylation, deamidation, and/or oxidation. Double hashtags denote a different combination of co-modifications within the same peptide (detailed in Supplementary Table S7). Error bars display the standard error of the mean from three (n = 3) independent reactions. (b) Schematic of hnRNPM showing nuclear localisation domain (NLS) RNA recognition motifs (RRM) and phospho-sites currently annotated in PhosphoSitePlus (Chk1 and PRP4) along with the novel MASTL sites identified here. Sequence alignments of the MASTL phosphorylation sites within hnRNPM (S-633 and S-637), compared to validated substrates Arpp19/ENSA and the possible MASTL motif identified from our SILAC screen (Fig. 2d).