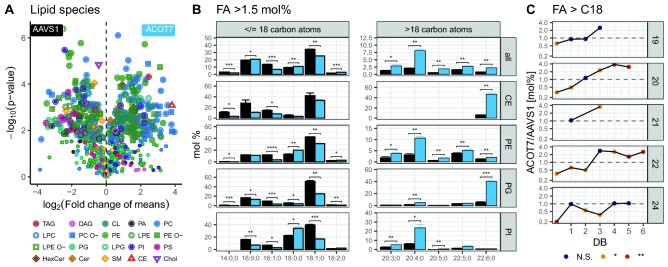
Figure 6.
ACOT7 knockout: (A) Volcano plot: Lipid species of the ACOT7 KO cell line (n = 3) and the AAVS1 control (n = 3) were compared. P-values of t-tests without correction for multiple testing are displayed on the y-axis, fold-changes of means are shown on the x-axis. Points with additional outlines indicate lipids significant after correction for multiple testing (Benjamini–Hochberg, 358 lipid species or 54.1% of all species). The shapes and colors of points indicate the lipid class of the species. (B) Fatty acid profile: greater than 1.5 mol% across CE, DAG, LPC, LPE, LPE O-, LPG, PA, PC, PC O-, PE, PE O-, PG, PI, PS, TAG lipid classes and individual profiles of CE, PE, PG, PI. Means are indicated by bar plots. Error bars correspond to standard deviations. p-values have been adjusted for the total number fatty residues (n = 50) or the total number fatty residues and lipid classes (n = 240) and are encoded as follows: * for q < 0.05, ** for q < 0.01, *** for q < 0.01, **** for q < 0.0001. (C) Fatty acid ratios: of FAs greater than 18 carbon atoms across CE, DAG, LPC, LPE, LPE O-, LPG, PA, PC, PC O-, PE, PE O-, PG, PI, PS, TAG lipid classes. Fatty acid length in carbon atoms is shown in panels and fatty acid double bonds on the x-axis. The ratio of the means ACOT7/AAVS1 [mol%] as depicted in (B) is shown on the y-axis, with the corresponding p-values corrected for multiple testing encoded in color. Only lipid species were used, which had at least 2 valid measurements in each of the two cell lines (n = 662). The ACOT7 KO clone G12 is displayed, which was created by the gene trap method.