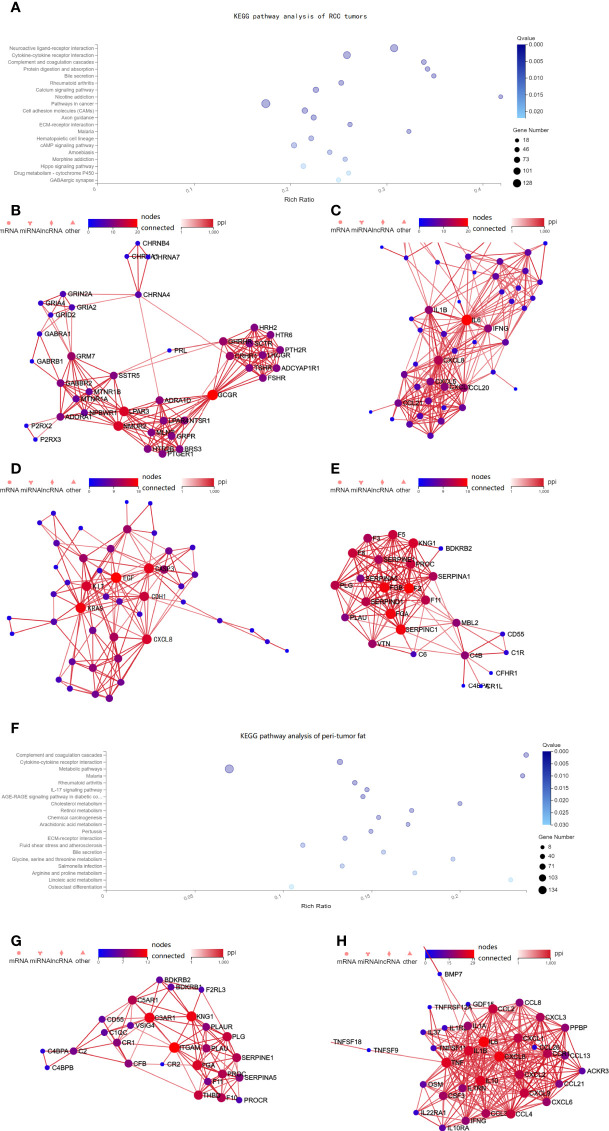
Figure 3.
Pathway alterations in the tumor tissue from the imaging genomics cohort. (A) KEGG enrichment analysis of annotated different expressed gene was performed in the imaging genomics cohort. Pathway analysis showed that the neuroactive ligand–receptor interaction, cytokine–cytokine receptor interaction, complement and coagulation cascades were altered in high FAIPTAT patients compared with low FAIPTAT patients. (B–E) The PPI networks were drawn using Cytoscape and key genes from those pathways such as GCGR, LPAR3, NMUR2, EGF, KIT, KRAS, CXCL8, IL-6, GCGR, FGA, and FGB were observed to be altered. (F) Pathway analysis showed that the complement and coagulation cascades, cytokine–cytokine receptor interaction, and multiple metabolic pathways were altered in high FAIPTAT patients compared with low FAIPTAT patients. (G–H) The PPI networks were drawn using Cytoscape, key genes such as ITGAM, FGA, and KNG1 were observed for the complement and coagulation cascades, while alterations of IL-6, TNF, CXCL8, IL-19, CXCL2, IL-1B, etc. were observed for the cytokine pathway.