Figure 5.
APTSTAT3-9R is effectively taken up by cells within the TME and inhibits STAT3 signaling in vemurafenib-resistant melanoma tumors
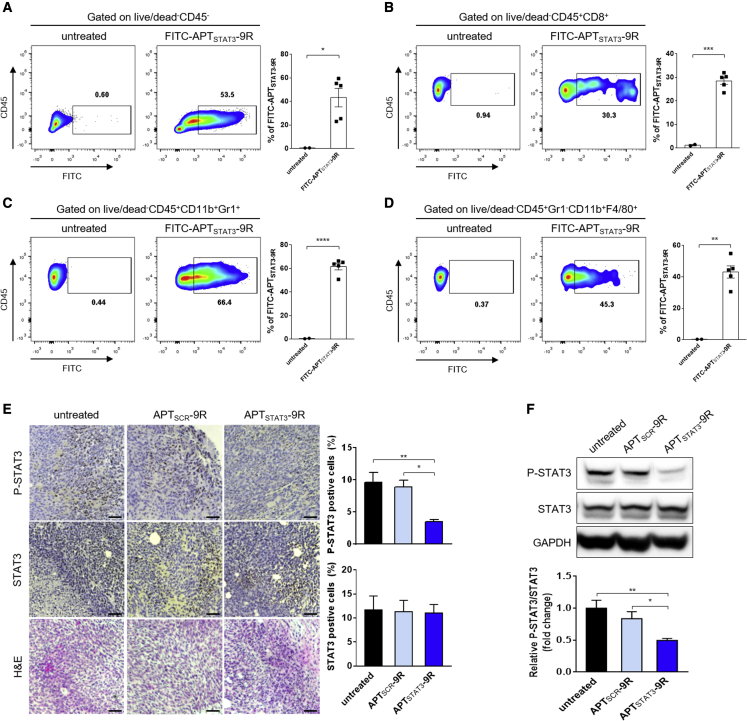
(A–D) Flow-cytometry analysis of TME cells positively stained with FITC-APTSTAT3-9R. When YUMM1.7R-derived tumors reached ∼300 mm3, mice were injected i.t. with FITC-APTSTAT3-9R (10 mg/kg), and tumors were harvested after 4 h for flow-cytometry analysis. Representative contour plots of (A) FITC-positive tumor/cancer-associated fibroblasts, obtained by gating on live/dead−CD45−; (B) MDSCs, obtained by gating on live/dead−CD45+CD11b+Gr1+; (C) TAMs, obtained by gating on live/dead−CD45+Gr1−CD11b+F4/80+; and (D) CD8+ T cells, obtained by gating on live/dead−CD45+CD8+. Data are presented as means ± SEM (n = 2 mice for the untreated group and n = 5 mice for FITC-APTSTAT3-9R-treated groups; ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; unpaired two-tailed Student’s t test). (E) Representative images showing immunohistochemical detection of P-STAT3 (Tyr705) and STAT3, and H&E staining in tissues of YUMM1.7R-derived tumors treated with each modality. When YUMM1.7R-derived tumors reached ∼100 mm3 (day 6), mice were injected i.t. with APTSCR-9R or APTSTAT3-9R (10 mg/kg) every other day for a total of four injections. Tumor sections were obtained on day 14. Scale bar, 100 μm. The bar graph shows quantification of P-STAT3 (Tyr705) and STAT3-positive cells. Data are presented as means ± SEM (n = 4 mice/group; ∗∗p < 0.01, ∗p < 0.05 versus untreated; one-way ANOVA followed by Sidak’s post hoc test). (F) Expression of P-STAT3 (Tyr705), STAT3, and GAPDH protein in YUMM1.7R-derived tumor tissues was determined by western blot analysis of lysates of tumors treated with either APTSCR-9R or APTSTAT3-9R as indicated in (A). Data are presented as means ± SEM (n = 4 mice/group; ∗∗p < 0.01, ∗p < 0.05 versus untreated; one-way ANOVA followed by Sidak’s post hoc test).