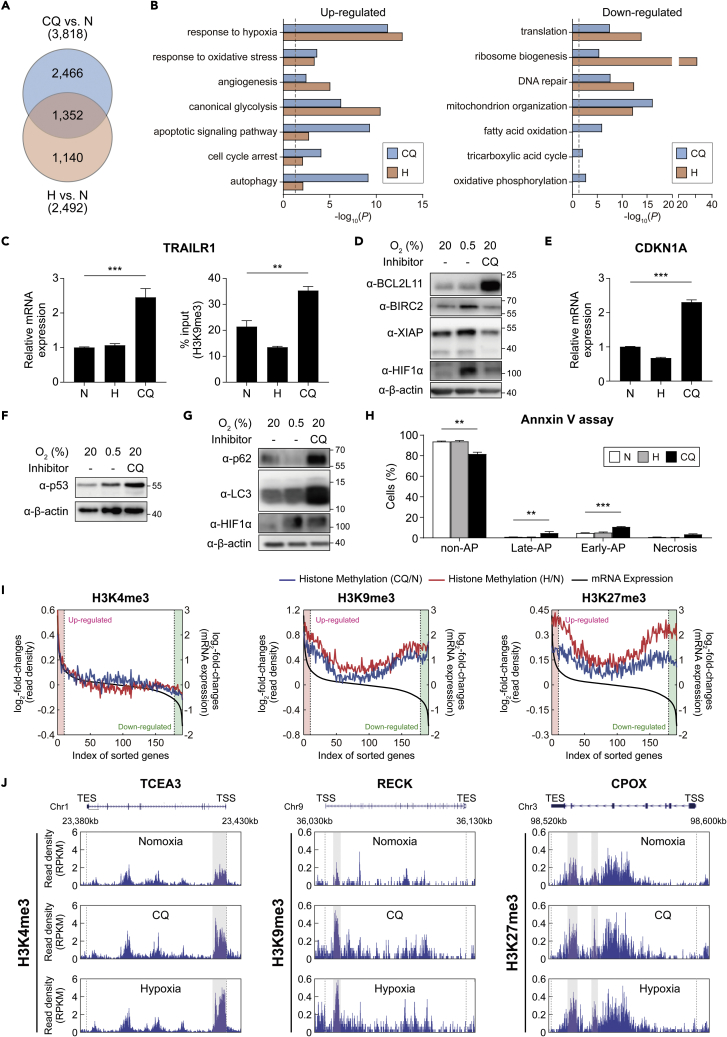
Figure 3.
CQ affects the expression of target genes and their associated cellular processes
(A) Comparison of the DEGs in hADSCs after CQ treatment and hypoxia. The numbers in the parentheses denote the total number of DEGs by CQ or hypoxia.
(B) GOBPs represented by the upregulated (left) and downregulated (right) genes by CQ or hypoxia. x axis, -log10(p), where p is the enrichment p value obtained using DAVID software. Dotted line, the cutoff value used to select the enriched GOBPs.
(C) Relative mRNA expression levels (left) and H3K9me3 levels (right) of the TRAILR1 gene in hADSCs, as measured using qRT-PCR and ChIP-PCR analyses, respectively, under N, H, or CQ. Data are shown as the mean ± SEM (n = 6 per condition).
(D) Representative western blot images showing the amounts of pro-apoptotic (BCL2L11) and anti-apoptotic (BIRC2 and XIAP) markers in hADSCs under N, H, or CQ (50 μM, 72 h).
(E and F) Relative mRNA expression levels of CDK1A. Western blot of p53 in hADSCs under N, H, or CQ. Data are shown as the mean ± SEM (n = 4 per condition). Statistical differences were assessed using a two-tailed paired Student’s t-test. ∗∗∗p < 0.001.
(G) Representative western blot images showing the amounts of an autophagy marker (p62/SQSTM1), LC3B, in hADSCs under N, H, and CQ (50 μM, 72 h).
(H) Percentages of non-apoptotic (AP), early-apoptotic, late-apoptotic, and necrotic hADSCs as measured via flow cytometry after annexin V/PI staining under normoxia (N), hypoxia (H) (0.5% O2, 48 h), or after treatment with CQ (CQ) (50 μM, 16 h). Data are presented as the mean ± SEM (n = 5 per condition).
(I) Correlation of log2-fold-changes in histone modifications with gene expression in individual histone methylations. The expressed genes were sorted using their log2-fold-changes and binned such that each bin includes 100 genes. For each type of histone methylation, the averaged log2-fold-changes of the RPKMs of the histone modification (y1-axis) and the averaged log2-fold-changes of gene expression levels (y2-axis) in individual bins (x axis) were plotted. Black lines represent the averaged log2-fold-changes of gene expression. Blue and red lines represent the averaged log2-fold-changes of histone methylation levels by CQ and hypoxia, respectively. Upregulated and downregulated genes (red and green backgrounds, respectively) were defined as those with fold-changes >1.5.
(J) The aligned reads of three representative genes (TCEA3, RECK, and CPOX) for the three histone methylations. The regions that include the differentially methylated peaks for H3K4me3, H3K9me3, and H3K27me3 were indicated by gray backgrounds. ∗∗p < 0.01 and ∗∗∗p < 0.001 by one-way ANOVA with Tukey’s correction (C and H).