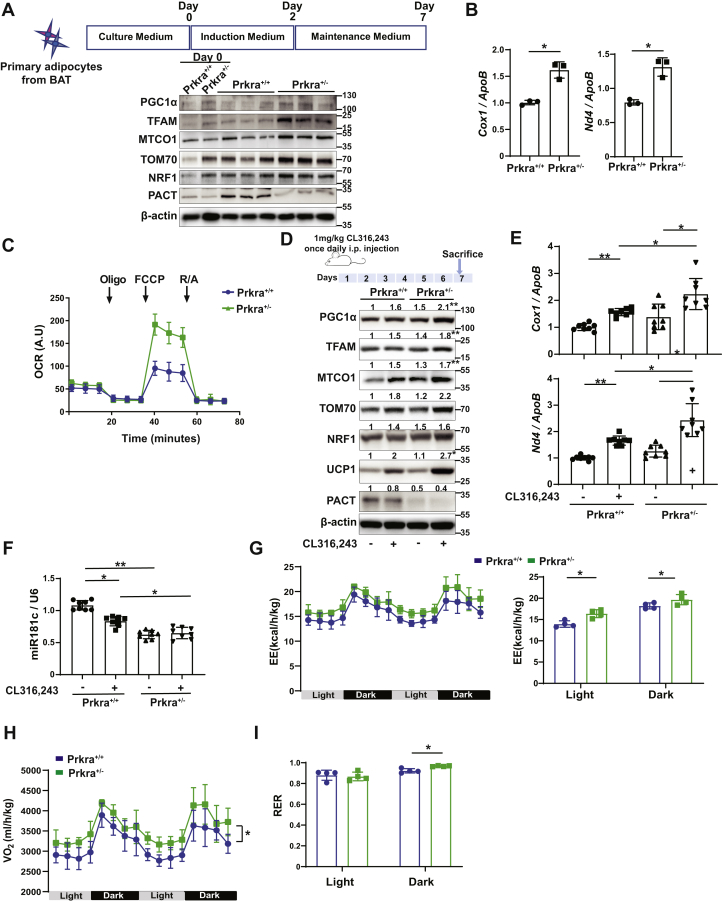
Figure 4.
Loss of PACT augments CL316,243-induced brown adipose mitobiogenesis.A, schematic representation of primary brown adipocyte differentiation protocol (upper panel). Mitochondrial protein levels were analyzed from protein lysates of undifferentiated (Day 0) or differentiated Prkra+/+ or Prkra+/− brown adipocytes by Western blotting using specific antibodies for PGC1α, TFAM, antibody cocktail against ETC proteins, TOM70, NRF1, and β-actin (n = 3). B, the genomic DNA obtained from differentiated brown adipocytes in (A) was analyzed by qRT-PCR for mtDNA (Cox1 or Nd4): nucDNA (ApoB) ratio (n = 3). C, mitochondrial respiration was analyzed by OCR in differentiated Prkra+/+ or Prkra+/− brown adipocytes (n = 6; data were normalized to mitochondrial mass quantified from total OXPHOS protein levels from same samples and represented as arbitrary units (A.U)). D–F, Prkra+/+ or Prkra+/− mice were injected with CL316,243 (1 mg/kg/day) for 6 days and sacrificed 24 h after the final injection (n = 8). D, BAT protein lysates were analyzed by Western blotting using specific antibodies for PGC1α, TFAM, antibody cocktail against ETC proteins, TOM70, NRF1, UCP1, and β-actin. E, mtDNA (Cox1 or Nd4): nucDNA (ApoB) ratio was analyzed by qRT-PCR from total genomic DNA. F, total BAT RNA extracts were analyzed by qRT-PCR for miR-181c and U6 levels. G–I, metabolic parameters of Prkra+/+ or Prkra+/− animals (n = 4): (G) energy expenditure (EE), (H) oxygen consumption (VO2), and (I) respiratory exchange ratio (RER). Protein expression was calculated relative to β-actin (for whole cell lysate) or Ponceau S. (for mitochondrial fraction) and depicted at the top of each blot. Data are mean ± SD. Unpaired t test with Welch’s correction or one-way ANOVA. ∗p ≤ 0.05, ∗∗p ≤ 0.01. BAT, brown adipose tissue; ETC, electron transfer chain; mtDNA, mitochondrial DNA; nucDNA, nuclear DNA; OCR, oxygen consumption rate.