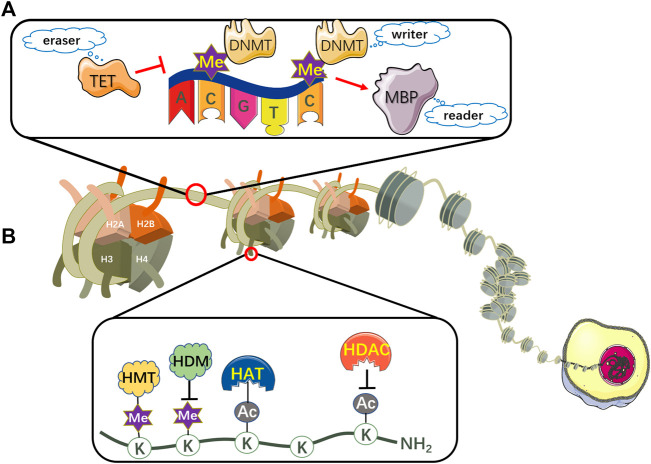
FIGURE 2.
The mechanism of DNA methylation and histone modifications. (A) DNA can be epigenetically modified by DNMT-based methylation. DNMTs are involved in catalyzing a methyl group to CpG dinucleotides, thus representing writers in epigenetic modifications. MBPs serve as readers to recognize and bind to the methylated CpG sites. TET proteins function as erasers to remove the epigenetic label of 5mC. (B) Nucleosomes comprise eight histone proteins including two copies of H2A, H2B, H3 and H4. Histone acetylation is maintained by the coordination of HATs and HDACs, which acetylate or deacetylate the lysine residues respectively in the N-terminal tails of histones protruding from the octamer. Histone lysine methylation is regulated by HMTs and HDMs, which transfer or remove highly processive methyl addition to the lysine. TET, ten-eleven translocation; DNMT, DNA methyltransferase; MBP, methyl-CpG binding protein; Me, methyl group; HAT, histone acetyltransferase; HDAC, histone deacetylase; HMT, histone methyltransferase; HDM, histone demethylase; K, lysine residue; AC, acetyl group.