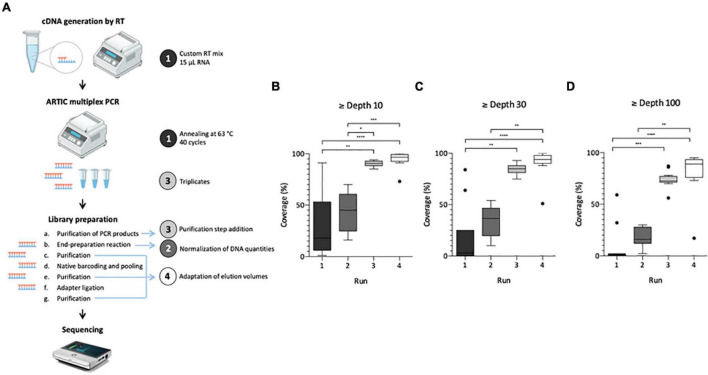
FIGURE 1.
Adaptation process of SARS-CoV-2 genome sequencing from wastewater samples. Left panel illustrates the protocol summary (A) with adaptation lines 1–4 depicted by a colored circle, same colors are used for box plot panels (B–D). Box Plot (Tukey whiskers) of SARS-CoV-2 genome coverage percentages obtained per run of sequencing on raw wastewater samples at depth 10 (B), 30 (C), and 100 (D) during the adaptation process of the ARTIC protocol. Adaptation lines were for Run 1: cDNA synthesis (15 μ1 RNA extract, random hexamers and SuperScript II reverse transcriptase), ARTIC multiplex PCR (annealing at 63°C, 40 cycles); Run 2: library preparation (normalization of initial DNA quantities); Run 3: ARTIC multiplex PCR (triplicates for each pool), library preparation (addition of an initial purification step of PCR products); Run 4: library preparation (addition of a purification step between the end-preparation and the barcoding reactions, adaptation of elution volumes to maximize recovery). Kruskal-Wallis test followed by Dunn’s multiple comparisons test were used to compare groups (****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05 and not significant if no indication on the plot) Panel A is adapted from Hourdel et al. (2020).