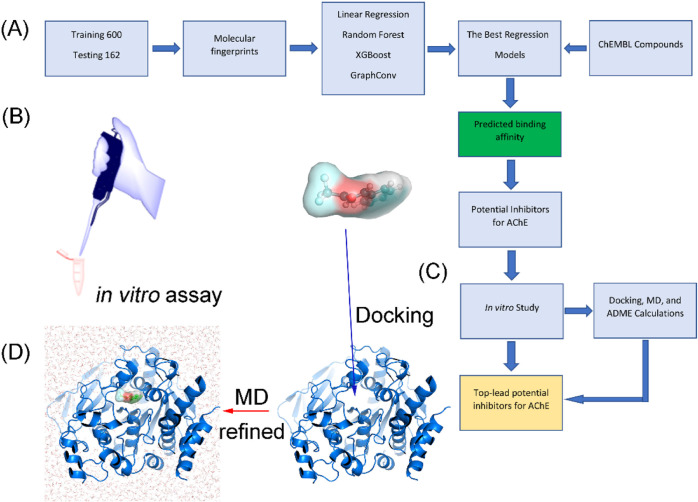
Figure 1.
Workflow for predicting potential inhibitors for AChE. (A) Investigation scheme was applied to estimate potential inhibitors for AChE using ML, atomistic calculations, and in vitro studies. (B) Refined investigation of the ML prediction via an in vitro enzyme assay. (C) Predicted potential inhibitors by the ML model were docked to the AChE active site via the modified AutoDock Vina.44 (D) AChE + trifluoromethylstyryl ketone complex was simulated using MD simulations to find the ligand-binding pose.