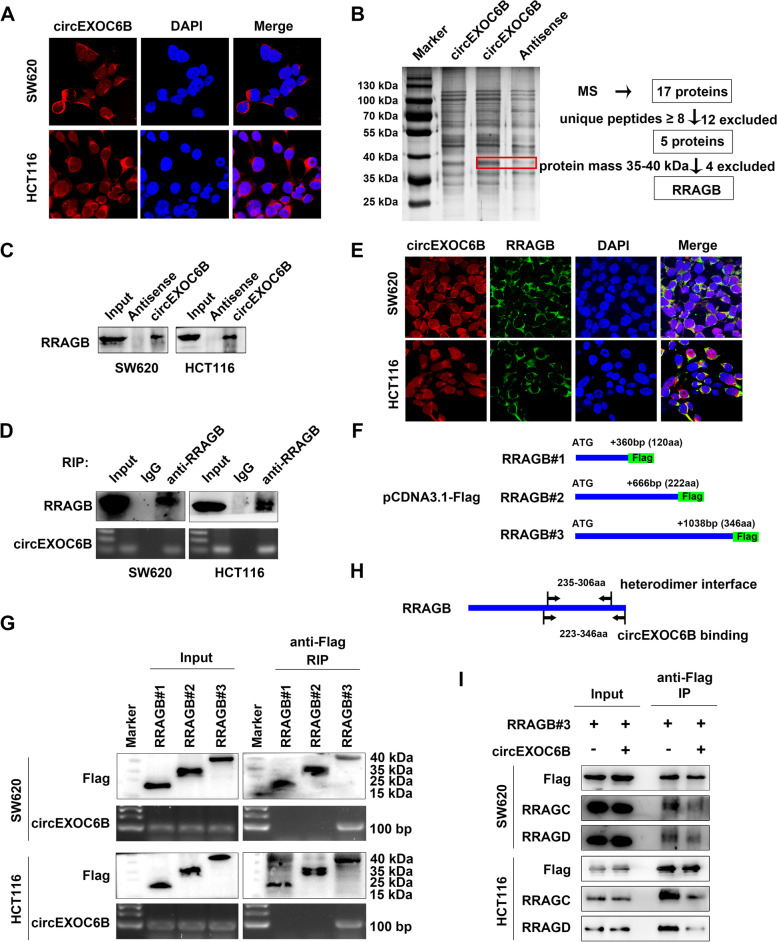
Fig. 3.
circEXOC6B binds to RRAGB to interfere with its heterodimer formation. A FISH showed that circEXOC6B was mainly resided in the cytoplasm of CRC cells; Red: circEXOC6B, Blue: nuclei. B RNA pull-down and mass spectrometry (MS) were performed to identify proteins binding to circEXOC6B. The red frame marked RRAGB. C RNA pull-down followed by western blot confirmed the binding of circEXOC6B and RRAGB. D RIP assay indicated that RRAGB could enrich circEXOC6B. E FISH and IF labeled circEXOC6B and RRAGB in CRC cells. F RRAGB expression vectors of different lengths (#1–3) were constructed using pCDNA3.1-Flag vector. G RIP assay showed that the 223–346-aa region of RRAGB was responsible for the interaction with circEXOC6B. H The 223–346-aa region of RRAGB contains the binding site for the heterodimerization of RRAGB with RRAGC/D (235–306-aa, NCBI gene database). I Co-IP demonstrated that the binding of RRAGC and RRAGD to RRAGB decreased with circEXOC6B overexpression