Figure 7.
Spatial interrogation of tissue microenvironmental determinants for retroviral transcription state
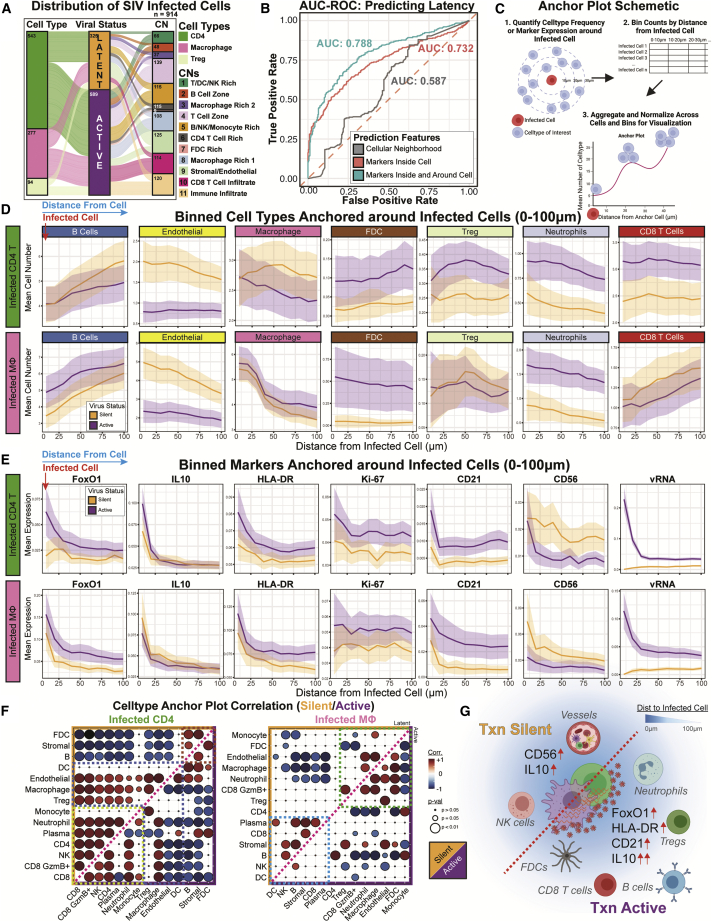
(A) An alluvial plot depicting the compositions of SIV-infected cell types (CD4+ T cells, n = 543; macrophages, n = 277; and Tregs, n = 94), their viral transcription status, and their associated CNs.
(B) Predictive performances of classifiers (1) CNs (gray, AUC = 0.587), (2) markers inside the infected cell (orange, AUC = 0.732), and (3) markers from a cell and its nearest neighbors (teal, AUC = 0.788). The dotted-red line indicates AUC of 0.5, as expected by chance.
(C) A schematic depicting how the anchor plots were calculated for the anchor plots in (D) and (E). In short, (1) mean cell type frequencies or marker expressions around each infected cell were tabulated, (2) these values were binned by their distance from the infected cell in 10-μm increments, and (3) data for all infected cells were aggregated and normalized for visualization.
(D and E) Anchor plots of (D) mean cell type quantifications and (E) mean marker expression around infected CD4+ T cells (top) or macrophages (bottom). Orange indicates transcriptionally silent cells, and purple indicates actively transcribing cells. The thick colored lines represent the means, and light regions around these lines depict the 95% confidence intervals.
(F) Heatmaps of Pearson’s correlations for cell type pairs for infected CD4+ T cells (left) and macrophages (right). Transcriptionally silent infection correlation heatmaps are represented in the top left (orange) and active infection correlation heatmaps are in the bottom right (purple). The sizes of the circles reflect the p values from the correlation test for association, and colors indicate degree of correlation.
(G) Schematic representing the tissue correlates and determinants of retroviral transcription status in retroviral reservoirs.
See also Figure S7.