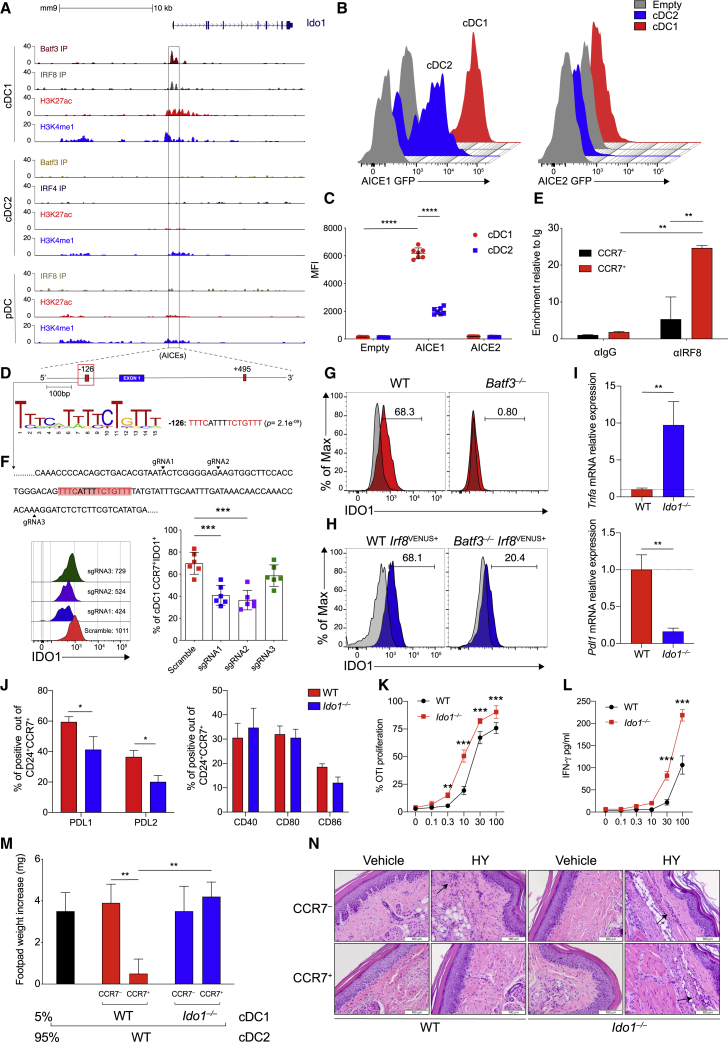
Figure 2.
IRF8 imprints constitutive IDO1 expression in cDC1
(A) ChIP-seq tracks display open chromatin areas and bindings of IRF8, BATF3, IRF4, H3K27ac, and H3K4me1 around Ido1 locus. Boxed areas at −126 bp or +495 bp from IDO1 TSS indicate regions assessed for enhancer activity.
(B and C) Flow cytometric analysis showing GFP-reporter activities (B) and quantification (C) in cDC1s and cDC2s expressing IDO1 −126 bp and + 495-bp enhancers (n = 7).
(D) FIMO analysis depicting p values of the two predicted AICEs (red boxes) in mouse Ido1 chr8: 25,694,453–25,713,138 (−126 bp from Ido1 TSS).
(E) IRF8 enrichment at the AICEs sequences of Ido1 promoter in sorted CCR7+ and CCR7− cDC1 (n = 2).
(F) IDO1 expression (MFI) in CCR7+ cDC1s differentiated from Rosa26Cas9−GFP/+ CD117hi BM progenitors expressing scramble RNA or sgRNA(s) (black arrowheads) targeting Ido1 −126 bp AICE1, as depicted in the single-color histograms (n = 3).
(G and H) IDO1 expression by flow in WT and Batf3−/− (H) and WT Irf8VENUS+ and Batf3−/−Irf8VENUS+ cDC1 (n = 3).
(I) Tnf and Pdl1 mRNA expression in sorted CCR7+ cDC1 of indicated phenotypes (n = 4).
(J) PDL1, PDL2, CD40, CD80, and CD86 expressions by flow in CCR7+ cDC1 (n = 3).
(K) Sorted WT or Ido1−/− cDC1 assayed for presentation to OT-I T cells in response to soluble OVA protein (n = 2).
(L) IFN-γ production in supernatants from (K) (n =2).
(M) Analysis of DTH is presented as change in footpad weight. n = 5 mice/group for (n = 2).
(N) H&E staining of mice footpad from mice in (M).
Data are shown as means ± SD. ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001, two-way ANOVA followed by Bonferroni multiple comparison test (C, E, F, J, K, and L) or Tukey’s multiple comparison test (M) and unpaired t test (I). Isotype control is shown as gray histogram. Please also see Figure S2.