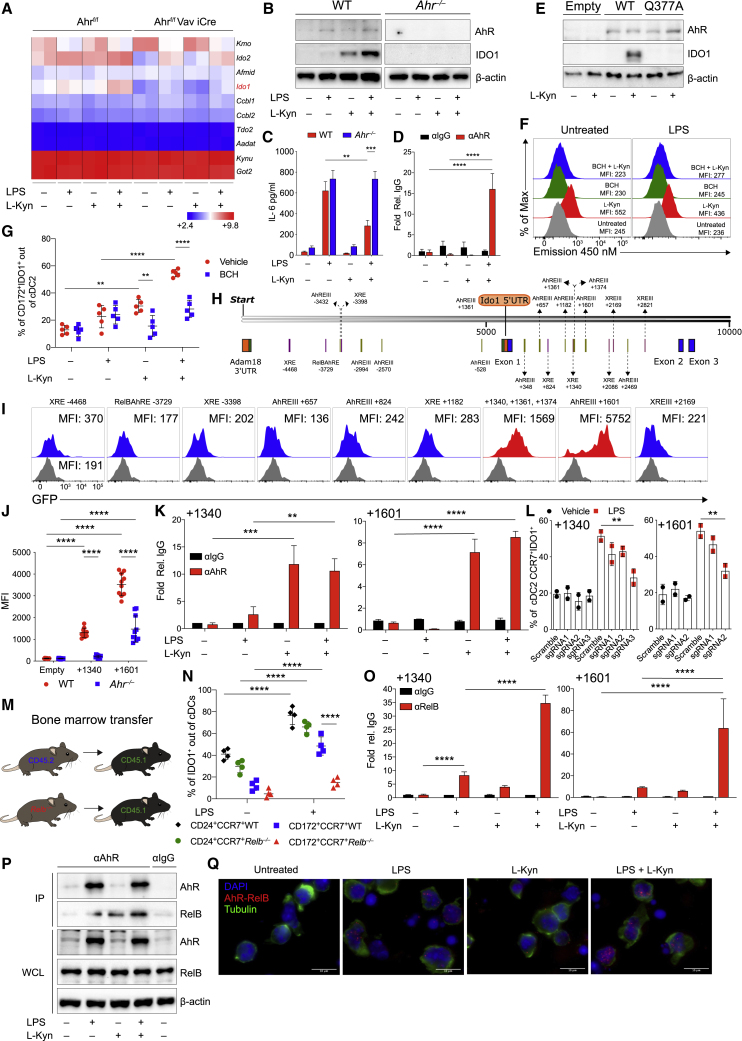
Figure 6.
AhR cooperates with RelB to induce IDO1 in isolated cDC2 treated with l-kynurenine
(A) Heatmap of tryptophan metabolic enzymes gene expression in cDC2 from AhRf/f mice versus Ahrf/fVav1 iCre mice treated as depicted. Bright blue (lowest) to bright red (highest).
(B) IDO1 immunoblot in cDC2 treated as shown for 48 h (n = 4).
(C) IL-6 analysis in supernatants of cDC2 treated as in (A) for 48 h (n = 4).
(D) AhR enrichment at the κB sequences of Il6 promoter by ChIP in sorted cDC2 untreated or treated overnight with LPS and conditioned for 2 h with l-kynurenine (n = 2).
(E) IDO1 immunoblot in purified cDC2 were transfected and treated as depicted for 48 h (n = 4).
(F) l-kynurenine uptake in purified cDC2. MFI is shown (n = 4).
(G) IDO1 expression by IS in cDC2 treated as (A) in the presence or absence of BCH for 48 h (n = 5).
(H) Predicted canonical AhR binding sites in the murine Ido1 promoter.
(I) MFI of GFP expression in pre-gated as Thy1.1+ cDC2 transduced with RV vector containing regions as described in (H). Gray histograms show empty reporter (n = 4).
(J) Quantification of the +1,340 bp and +1,601 bp Ido1 enhancer activity in WT and Ahr−/− cDC2s using retroviral reporters as in (I) (n = 3).
(K) ChIP-PCR analysis of AhR binding on +1,340 bp and +1,601 bp Ido1 enhancer elements in cDC2 treated as in (C) (n = 3).
(L) IDO1 expression by IS in gated CCR7+cDC2 derived from Rosa26Cas9−GFP/+ c-Kithi progenitors infected with RV expressing sgRNAs targeting +1,340 bp and +1,601 bp Ido1 enhancer elements treated as shown for 48 h (n = 2).
(M) Schematic representation of BM chimera model.
(N) IDO1 expression by IS in cDC1 and cDC2 from BMDC derived as in (M) and cultured as shown (n = 3).
(O) ChIP-PCR analysis of RelB binding on +1,340 bp and +1,601 bp Ido1 enhancer elements in cDC2 treated as in (C) (n = 3).
(P) AhR and RelB Immunoblot in purified cDC2 treated as in (K) where AhR was immunoprecipitated (n = 3).
(Q) AhR and RelB interaction in purified cDC2 treated as indicated by PLA. Red spots show a single AhR/RelB interaction. Scale bars, 10 μm (n = 3).
Data are shown as means ± SD. ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001, one-way (L) or two-way (C, D, G, J, K, N, and O) ANOVA followed by Bonferroni multiple comparison test. Dots represent a biological replicate (G and N). β-actin used as loading control (B, E, and P). Data are normalized to IgG control (D, K, and O). Please also see Figure S6.