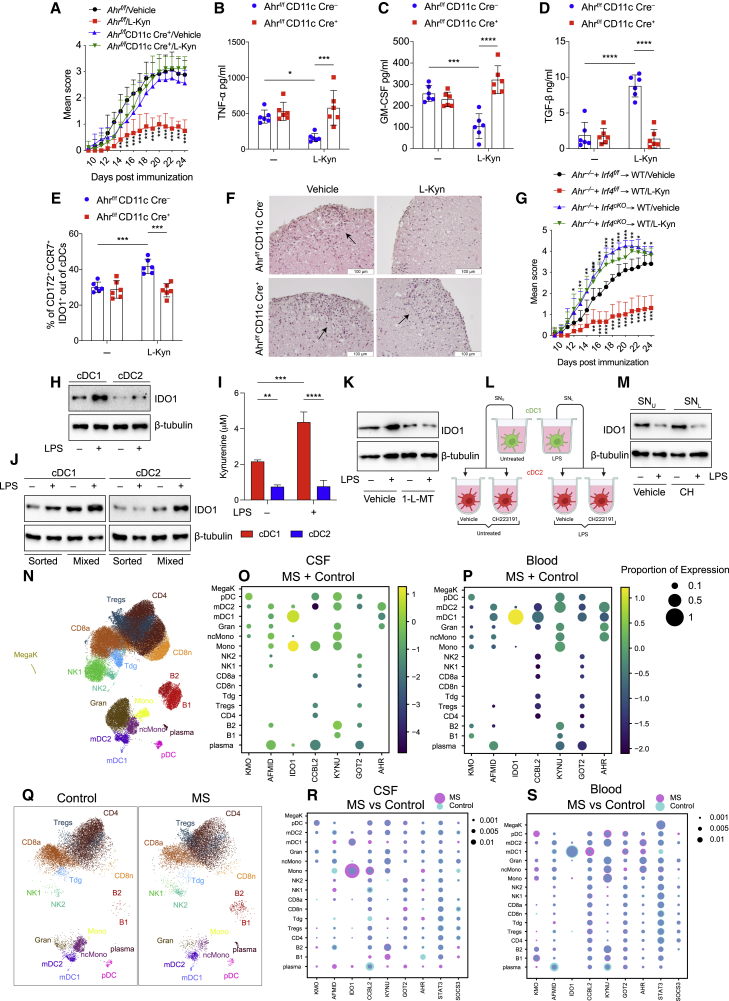
Figure 7.
l-kynurenine induces immune regulatory functions in EAE
(A) EAE score in Ahrf/fItgax Cre− or Ahrf/fItgax Cre+ mice treated with l-kyn or vehicle, from 11 to 22 days post immunization. 6 mice per group, n = 3.
(B–D) TNF-α (B), GM-CSF (C), and TGFβ (D) in plasma from EAE mice in (A).
(E) IDO1+CCR7+ cDC2 of cDCs by IS in cervical LNs from EAE mice treated as in (A).
(F) H&E staining of spinal cord sections from mice treated in (A). Scale bar, 100 μM.
(G) EAE score of mixed bone marrow chimeras with bone marrow of indicated genotypes. 9 mice per group, n = 2.
(H) IDO1 immunoblot in cDC1 and cDC2, sorted from human PBMCs, were treated as depicted for 36 h (n = 3).
(I) l-kynurenine content in supernatants from cells prepared as in (H) (n = 3).
(J) IDO1 immunoblot in purified human cDC1 and cDC2 cultured alone (sorted) or together (mixed), in trans-well plates, with or without LPS for 36 h (n = 3).
(K) IDO1 immunoblot in human cDC1 and cDC2 co-cultured in trans-well plates and treated as depicted for 36 h (n = 3).
(L) Schematic representation of human cDC2 treatment with cDC1 conditioned media. cDC1 untreated (SNU), LPS treated (SNL) for 36 h, CH2223191 (CH) cDC2 pre-treated for 2 h and incubated with cDC1 conditioned media for 36 h.
(M) IDO1 immunoblot in cDC2 treated as in (L) (n = 3).
(N) UMAP plot representing 17 color-coded cell clusters identified in merged single-cell transcriptomes of blood (42,969) and CSF (22,357) cells from control (n = 4) and multiple sclerosis (MS; n = 4) patients.
(O and P) Dot plot depicting selected genes of tryptophan metabolic pathway in cell clusters of CFS (O) and blood (P).
(Q) Comparative UMAP plots depicting only CSF cells from control (12,705 cells, left plot) and MS (9,652 cells, right plot) donors.
(R and S) Dot plot depicting selected genes differentially expressed in at least one cluster of MS cells compared with controls in CSF (R) or blood (S). Purple indicates higher and turquoise indicates lower expression in MS, respectively.
Data are shown as means ± SD. ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001, two-way ANOVA followed by Bonferroni multiple comparison test (A–E, G, and I). Dots represent a biological replicate (B–E). β-tubulin used as loading control (H, J, K, and M). Dot size encodes percentage of cells expressing the gene (O, P, R, and S). Please also see Figure S7.