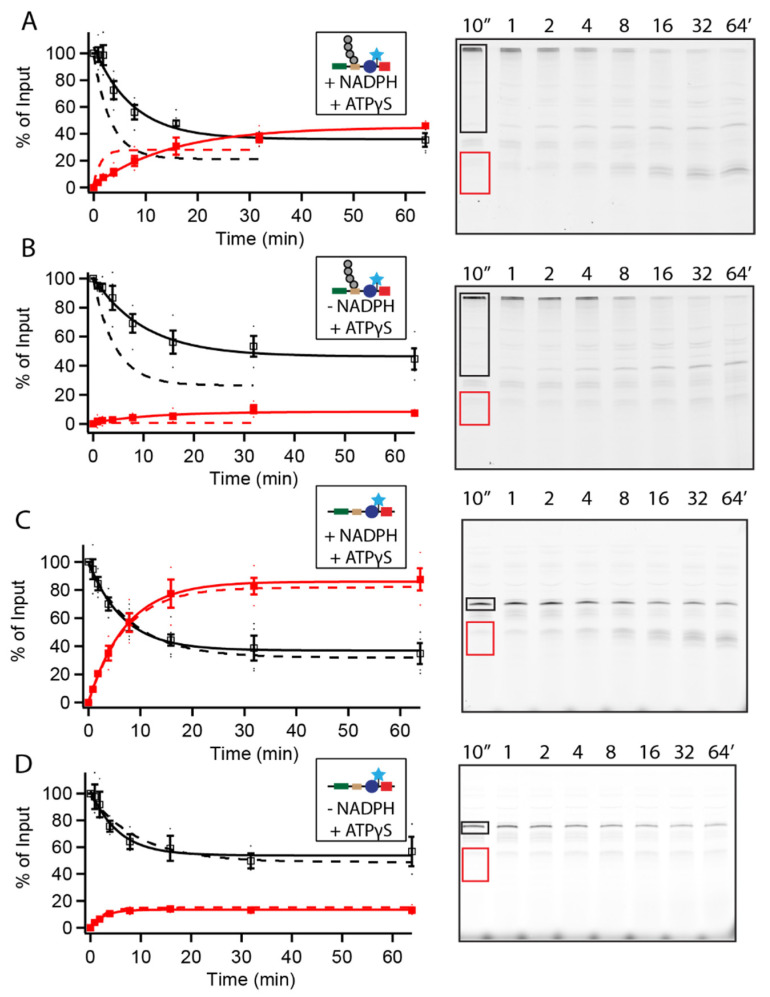
Figure 2.
Degradation of UbID substrates is ATP-independent. (A–D) Degradation of 20 nM Rpn41–80-PPXY-Barnase∆KL89G-C-DHFRk∆C by 100 nM WT proteasome in the presence of ATP-γS. Example gels show full-length substrate, (A,B) ubiquitinated or (C,D) nonubiquitinated, outlined in black, and DHFR fragment outlined in red. Full-length (open squares) and DHFR fragment (closed squares) are shown as a percentage of total full-length present at the beginning of the reaction; full length is quantified as the sum of ubiquitinated and non-ubiquitinated substrate so any deubiquitination is not misinterpreted as degradation. Dots are the results from individual experiments and error bars represent the SEM of 3–4 experiments. Dashed lines are fits in the absence of ATP-γS from Figure 1.