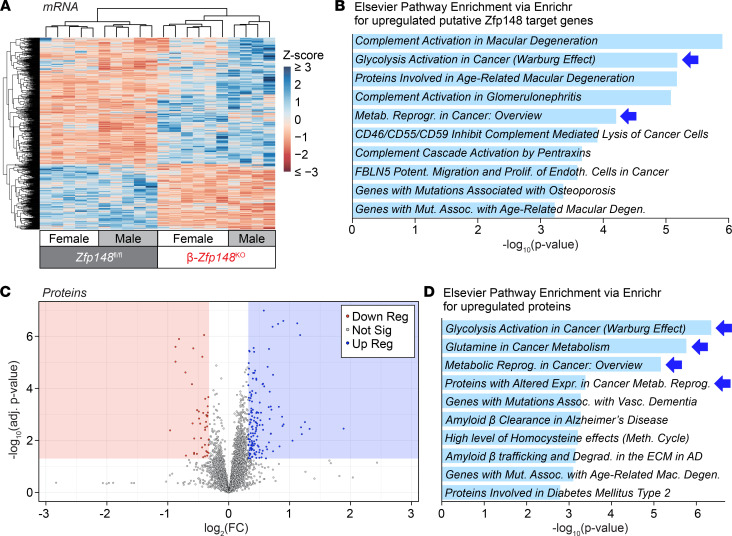
Figure 3. Altered expression of metabolic genes and proteins in β-Zfp148KO and control islets.
(A) Heat map of z-scores for significantly DE genes in RNA-Seq from islets of 10 WD-fed Zfp148fl/fl control and β-Zfp148KO mice, as determined using EBSeq. (B) Elsevier database pathway enrichment (from Enrichr; refs. 46 and 47) for upregulated, putative, direct DE targets of Zfp148, based on analysis in Supplemental Figure 4. (C) Volcano plot of proteins significantly increased or decreased in islets of 6 WD-fed β-Zfp148KO vs. 10 WD-fed Zfp148fl/fl control mice. (D) Elsevier database pathway enrichment for proteins significantly increased in β-Zfp148KO islets. For pathway enrichments, terms in italics indicate enrichment beyond the false discovery cutoff (adjusted P < 0.05). Corresponding genes for the indicated terms are listed in extended data in Dryad (46). (B and D) Blue arrows indicate related enrichment terms from both RNA and protein data sets. AD, Alzheimer disease; adj, adjusted; assoc, associated; degen, degeneration; degrad, degradation; ECM, extracellular matrix; endoth, endothelial; expr, expression; FC, fold change; mac, macular; metab, metabolic; meth, methionine; mut, mutations; potent, potentiates; prolif, proliferation; reprog, reprogramming; sig, significant; vasc, vascular.