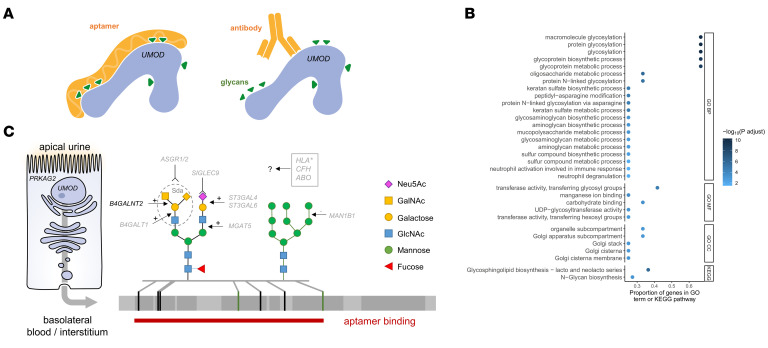
Figure 4. Biological context of genes associated with circulating uromodulin and conceptual model.
(A) Schematic of antibody- and aptamer-based measurement of circulating uromodulin. (B) Dot plot shows Gene Ontology (GO) terms — grouped into 3 categories (BP, biological process; MF, molecular function; CC, cellular component) — and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched for uromodulin-associated genes from the aptamer assay on the y axis. The x axis shows the proportion of the genes in the corresponding GO term or KEGG pathway. Only terms and pathways with more than 2 uromodulin-associated genes are displayed. The color intensity of the dots scales with the –log10(Benjamini-Hochberg–adjusted P value). (C) Conceptual model placing the most likely causal genes associated with circulating uromodulin into their biological context. Loci detected with the aptamer assay predominantly affect differential synthesis or recognition of glycan marks present on uromodulin. Gal, galactose; Glc, glucose; NAc, N-acetylgalactosamine; Neu5Ac, N-acetylneuraminic acid.