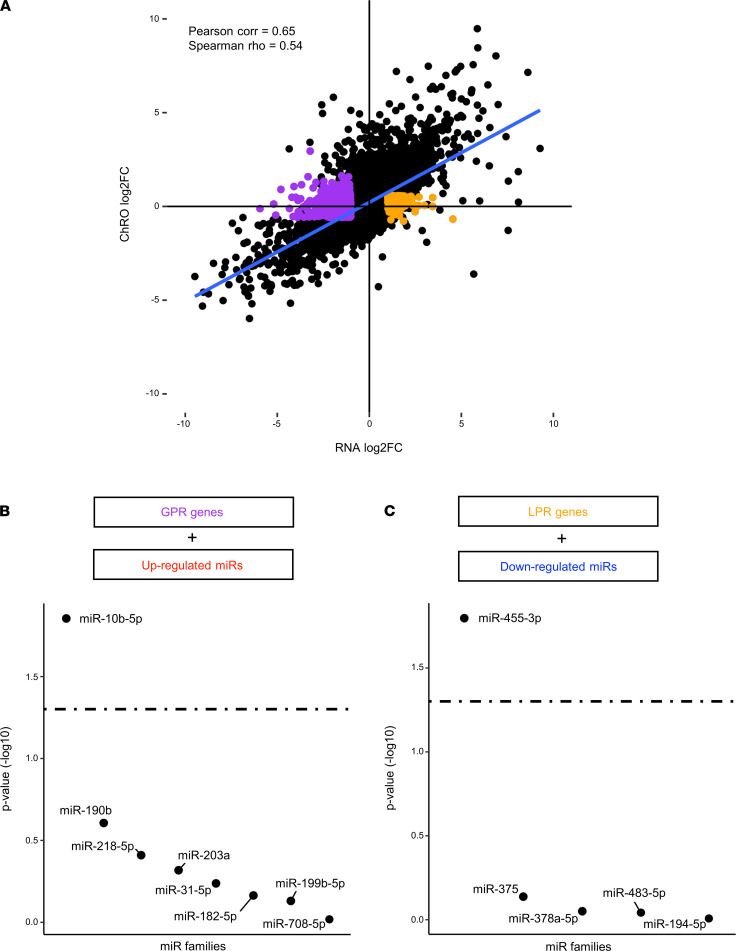
Figure 7. miR-10b and miR-455 are master regulators of gene expression in FLC.
(A) Scatterplot showing the log2 FC of RNA-Seq normalized reads on the x axis and the log2 FC of ChRO-Seq normalized reads on the y axis for genes in FLC relative to NML. Those genes subject primarily to gain of PTR (normalized reads > 1000, RNA-Seq log2 FC < 1, RNA-Seq Padj < 0.05, ChRO-Seq log2FC < ±0.59, ChRO-Seq Padj > 0.2) are highlighted in purple, and genes subject primarily to loss of PTR (normalized reads > 1000, RNA-Seq log2 FC > 1, RNA-Seq Padj < 0.05, ChRO-Seq log2FC < ±0.59, ChRO-Seq Padj > 0.2) are highlighted in orange. (B) Ranked –log10 (P value) of miRhub simulation results. Gain of PTR genes were examined for enrichment of binding sites for microRNAs upregulated in FLC (only those microRNAs with predictions in TargetScan included). The dashed line represents P = 0.05. (C) Ranked –log10 (P value) of miRhub simulation results. Loss of PTR genes was examined for enrichment of binding sites for microRNAs downregulated in FLC (only those microRNAs with predictions in TargetScan included). The dashed line represents P = 0.05.