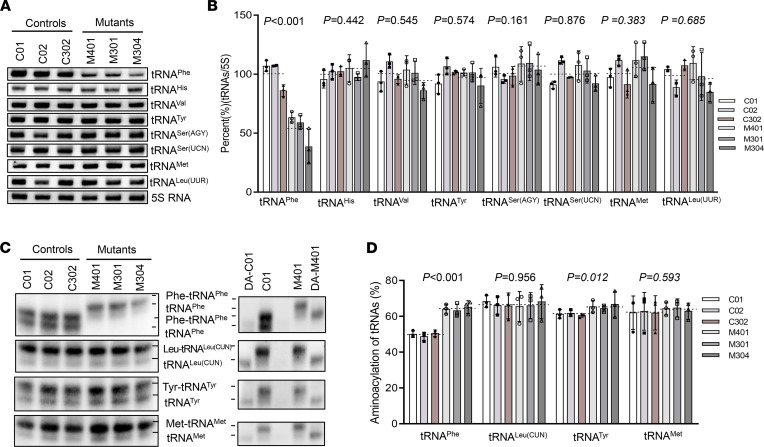
Figure 5. Northern blot analysis of mt-tRNA under denaturing conditions.
(A) Northern blot analysis of mt-tRNA under denaturing conditions. DIG-labeled oligodeoxynucleotide probes specific for the mt-tRNAPhe, mt-tRNAHis, mt-tRNAVal, mt-tRNASer(AGY), mt-tRNAMet, mt-tRNALeu(UUR), mt-tRNASer (UCN), mt-tRNATyr, and 5S rRNA as a loading control. (B) Quantification of mt-tRNAPhe levels. (C) Aminoacylation assays. RNA was hybridized with specific probes of mt-tRNAPhe, mt-tRNALeu(CUN), and mt-tRNATyr, and deacylation treatment was performed in parallel to distinguish charged and uncharged tRNAs. DA, deacylated. (D) The proportion of aminoacylated mt-tRNAPhe in control and mutant cell lines. The calculations were based on 3 independent experiments. The data are expressed as the means ± SD (n = 3). Student’s t test was performed to determine statistically significant differences.