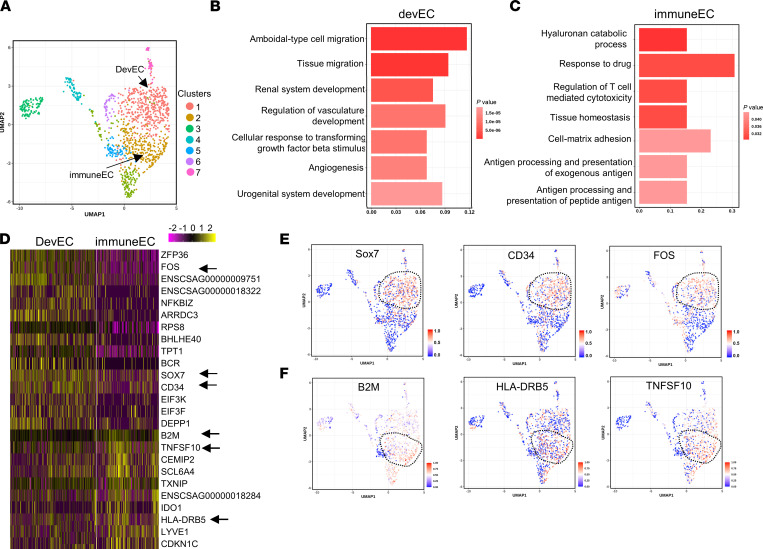
Figure 7. Analysis of lung ECs obtained from nonhuman primates infected with SARS-CoV-2.
African green monkeys were inoculated with 2.6 × 106 TCID50 (50% tissue culture infectious dose) live SARS-CoV-2 (nCoV-WA1-2020 MN985325.1). The scRNA-Seq was performed using harvested lung ECs from 4 nonhuman primates at day 10 after inoculation (recovered from SARS-CoV-2 infection). (A) UMAP of 3253 individual lung ECs; different colors indicate distinct clusters (immuneEC, immune EC subpopulation; devEC, developmental EC subpopulation). (B and C) The GO terms of devEC (B) and immuneEC (C) indicate the enriched biological processes in developmental signature (B) and immune signature (C). P values are indicated on the left in a color scale. X axis represents the percentage of differentially expressed genes in each GO term. (D) Heatmap of the most differentially expressed genes in lung ECs from devEC and immuneEC subpopulations. The color bars indicate gene expression level in log2 scale. Arrows highlight genes shown in E and F. UMAP of visualization of expression levels of the representative marker genes in devECs (E) and immuneECs (F). The color bars in UMAP indicate gene expression level using a log2 scale.